PhyloP regulatory elements
Briana Mittleman
12/21/2020
Last updated: 2021-01-18
Checks: 7 0
Knit directory: Comparative_APA/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190902) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version b769a17. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: code/chimp_log/
Ignored: code/human_log/
Ignored: data/.DS_Store
Ignored: data/TrialFiltersMeta.txt.sb-9845453e-R58Y0Q/
Ignored: data/mediation_prot/
Ignored: data/metadata_HCpanel.txt.sb-284518db-RGf0kd/
Ignored: data/metadata_HCpanel.txt.sb-a5794dd2-i594qs/
Ignored: output/.DS_Store
Untracked files:
Untracked: ._.DS_Store
Untracked: Chimp/
Untracked: GEO/
Untracked: Human/
Untracked: analysis/ALUelements.Rmd
Untracked: analysis/AREstabilityScores.Rmd
Untracked: analysis/AllLoc_effectSizeCor.Rmd
Untracked: analysis/Conservation_bydAPAset.Rmd
Untracked: analysis/CrossChimpThreePrime.Rmd
Untracked: analysis/DiffTransProtvsExpression.Rmd
Untracked: analysis/DiffUsedUTR.Rmd
Untracked: analysis/DirSelectionGenes.Rmd
Untracked: analysis/GvizPlots.Rmd
Untracked: analysis/HandC.TvN
Untracked: analysis/PhenotypeOverlap10.Rmd
Untracked: analysis/ResultsNoUnlifted.md
Untracked: analysis/SuppTables.Rmd
Untracked: analysis/annotationBias.Rmd
Untracked: analysis/assessReadQual.Rmd
Untracked: analysis/dUsed_humanchimp.Rmd
Untracked: analysis/diffExpressionPantro6.Rmd
Untracked: analysis/isoformdivandexp.Rmd
Untracked: analysis/nDom.Rmd
Untracked: code/._AlignmentScores.sh
Untracked: code/._BothFCMM.sh
Untracked: code/._BothFCMMPrim.sh
Untracked: code/._BothFCnewOInclusive.sh
Untracked: code/._ChimpStarMM2.sh
Untracked: code/._ClassifyLeafviz.sh
Untracked: code/._ClosestorthoEx.sh
Untracked: code/._Config_chimp.yaml
Untracked: code/._Config_chimp_full.yaml
Untracked: code/._Config_human.yaml
Untracked: code/._ConvertJunc2Bed.sh
Untracked: code/._CountNucleotides.py
Untracked: code/._CrossMapChimpRNA.sh
Untracked: code/._CrossMapThreeprime.sh
Untracked: code/._DiffSplice.sh
Untracked: code/._DiffSplicePlots.sh
Untracked: code/._DiffSplicePlots_gencode.sh
Untracked: code/._DiffSplice_gencode.sh
Untracked: code/._DiffSplice_removebad.sh
Untracked: code/._Filter255MM.sh
Untracked: code/._FilterPrimSec.sh
Untracked: code/._FindIntronForDomPAS.sh
Untracked: code/._FindIntronForDomPAS_DF.sh
Untracked: code/._GetMAPQscore.py
Untracked: code/._GetSecondaryMap.py
Untracked: code/._Lift5perPAS.sh
Untracked: code/._LiftFinalChimpJunc2Human.sh
Untracked: code/._LiftOrthoPAS2chimp.sh
Untracked: code/._MapBadSamples.sh
Untracked: code/._MismatchNumbers.sh
Untracked: code/._PAS_ATTAAA.sh
Untracked: code/._PAS_ATTAAA_df.sh
Untracked: code/._PAS_seqExpanded.sh
Untracked: code/._PASsequences.sh
Untracked: code/._PASsequences_DF.sh
Untracked: code/._PlotNuclearUsagebySpecies.R
Untracked: code/._PlotNuclearUsagebySpecies_DF.R
Untracked: code/._QuantMergedClusters.sh
Untracked: code/._RNATranscriptDTplot.sh
Untracked: code/._ReverseLiftFilter.R
Untracked: code/._RunFixLeafCluster.sh
Untracked: code/._RunNegMCMediation.sh
Untracked: code/._RunNegMCMediationDF.sh
Untracked: code/._RunPosMCMediationDF.err
Untracked: code/._RunPosMCMediationDF.sh
Untracked: code/._SAF2Bed.py
Untracked: code/._Snakefile
Untracked: code/._SnakefilePAS
Untracked: code/._SnakefilePASfilt
Untracked: code/._SortIndexBadSamples.sh
Untracked: code/._StarMM2.sh
Untracked: code/._TestFC.sh
Untracked: code/._assignPeak2Intronicregion
Untracked: code/._assignPeak2Intronicregion.sh
Untracked: code/._bed215upbed.py
Untracked: code/._bed2Bedbothstrand.py
Untracked: code/._bed2SAF_gen.py
Untracked: code/._buildIndecpantro5
Untracked: code/._buildIndecpantro5.sh
Untracked: code/._buildLeafviz.sh
Untracked: code/._buildLeafviz_leadAnno.sh
Untracked: code/._buildStarIndex.sh
Untracked: code/._chimpChromprder.sh
Untracked: code/._chimpMultiCov.sh
Untracked: code/._chimpMultiCov255.sh
Untracked: code/._chimpMultiCovInclusive.sh
Untracked: code/._chooseSignalSite.py
Untracked: code/._cleanbed2saf.py
Untracked: code/._cluster.json
Untracked: code/._cluster2bed.py
Untracked: code/._clusterLiftReverse.sh
Untracked: code/._clusterLiftReverse_removebad.sh
Untracked: code/._clusterLiftprimary.sh
Untracked: code/._clusterLiftprimary_removebad.sh
Untracked: code/._converBam2Junc.sh
Untracked: code/._converBam2Junc_removeBad.sh
Untracked: code/._extraSnakefiltpas
Untracked: code/._extractPhyloReg.py
Untracked: code/._extractPhyloRegGene.py
Untracked: code/._extractPhylopGeneral.ph
Untracked: code/._extractPhylopGeneral.py
Untracked: code/._extractPhylopReg200down.py
Untracked: code/._extractPhylopReg200up.py
Untracked: code/._filter5percPAS.py
Untracked: code/._filterNumChroms.py
Untracked: code/._filterPASforMP.py
Untracked: code/._filterPostLift.py
Untracked: code/._filterPrimaryread.py
Untracked: code/._filterSecondaryread.py
Untracked: code/._fixExonFC.py
Untracked: code/._fixFCheadforExp.py
Untracked: code/._fixLeafCluster.py
Untracked: code/._fixLiftedJunc.py
Untracked: code/._fixUTRexonanno.py
Untracked: code/._formathg38Anno.py
Untracked: code/._formatpantro6Anno.py
Untracked: code/._getRNAseqMapStats.sh
Untracked: code/._hg19MapStats.sh
Untracked: code/._humanChromorder.sh
Untracked: code/._humanMultiCov.sh
Untracked: code/._humanMultiCov255.sh
Untracked: code/._humanMultiCov_inclusive.sh
Untracked: code/._intersectLiftedPAS.sh
Untracked: code/._liftJunctionFiles.sh
Untracked: code/._liftPAS19to38.sh
Untracked: code/._liftedchimpJunc2human.sh
Untracked: code/._makeNuclearDapaplots.sh
Untracked: code/._makeNuclearDapaplots_DF.sh
Untracked: code/._makeSamplyGroupsHuman_TvN.py
Untracked: code/._mapRNAseqhg19.sh
Untracked: code/._mapRNAseqhg19_newPipeline.sh
Untracked: code/._maphg19.sh
Untracked: code/._maphg19_subjunc.sh
Untracked: code/._mediation_test.R
Untracked: code/._mergeChimp3prime_inhg38.sh
Untracked: code/._mergeandBWRNAseq.sh
Untracked: code/._mergedBam2BW.sh
Untracked: code/._nameClusters.py
Untracked: code/._negativeMediation_montecarlo.R
Untracked: code/._negativeMediation_montecarloDF.R
Untracked: code/._numMultimap.py
Untracked: code/._overlapMMandOrthoexon.sh
Untracked: code/._overlapPASandOrthoexon.sh
Untracked: code/._overlapapaQTLPAS.sh
Untracked: code/._parseHg38.py
Untracked: code/._postiveMediation_montecarlo_DF.R
Untracked: code/._prepareCleanLiftedFC_5perc4LC.py
Untracked: code/._prepareLeafvizAnno.sh
Untracked: code/._preparePAS4lift.py
Untracked: code/._primaryLift.sh
Untracked: code/._processhg38exons.py
Untracked: code/._quantJunc.sh
Untracked: code/._quantJunc_TEST.sh
Untracked: code/._quantJunc_removeBad.sh
Untracked: code/._quantLiftedPASPrimary.sh
Untracked: code/._quantMerged_seperatly.sh
Untracked: code/._recLiftchim2human.sh
Untracked: code/._revLiftPAShg38to19.sh
Untracked: code/._reverseLift.sh
Untracked: code/._runCheckReverseLift.sh
Untracked: code/._runChimpDiffIso.sh
Untracked: code/._runCountNucleotides.sh
Untracked: code/._runFilterNumChroms.sh
Untracked: code/._runHumanDiffIso.sh
Untracked: code/._runNuclearDiffIso_DF.sh
Untracked: code/._runNuclearDifffIso.sh
Untracked: code/._runTotalDiffIso.sh
Untracked: code/._run_chimpverifybam.sh
Untracked: code/._run_verifyBam.sh
Untracked: code/._snakemake.batch
Untracked: code/._snakemakePAS.batch
Untracked: code/._snakemakePASchimp.batch
Untracked: code/._snakemakePAShuman.batch
Untracked: code/._snakemake_chimp.batch
Untracked: code/._snakemake_human.batch
Untracked: code/._snakemakefiltPAS.batch
Untracked: code/._snakemakefiltPAS_chimp
Untracked: code/._snakemakefiltPAS_chimp.sh
Untracked: code/._snakemakefiltPAS_human.sh
Untracked: code/._spliceSite2Fasta.py
Untracked: code/._submit-snakemake-chimp.sh
Untracked: code/._submit-snakemake-human.sh
Untracked: code/._submit-snakemakePAS-chimp.sh
Untracked: code/._submit-snakemakePAS-human.sh
Untracked: code/._submit-snakemakefiltPAS-chimp.sh
Untracked: code/._submit-snakemakefiltPAS-human.sh
Untracked: code/._subset_diffisopheno_Nuclear_HvC.py
Untracked: code/._subset_diffisopheno_Nuclear_HvC_DF.py
Untracked: code/._subset_diffisopheno_Total_HvC.py
Untracked: code/._threeprimeOrthoFC.sh
Untracked: code/._transcriptDTplotsNuclear.sh
Untracked: code/._verifyBam4973.sh
Untracked: code/._verifyBam4973inHuman.sh
Untracked: code/._wrap_chimpverifybam.sh
Untracked: code/._wrap_verifyBam.sh
Untracked: code/._writeMergecode.py
Untracked: code/.snakemake/
Untracked: code/ALLPAS_sequenceDF.err
Untracked: code/ALLPAS_sequenceDF.out
Untracked: code/AlignmentScores.err
Untracked: code/AlignmentScores.out
Untracked: code/AlignmentScores.sh
Untracked: code/BothFCMM.err
Untracked: code/BothFCMM.out
Untracked: code/BothFCMM.sh
Untracked: code/BothFCMMPrim.err
Untracked: code/BothFCMMPrim.out
Untracked: code/BothFCMMPrim.sh
Untracked: code/BothFCnewOInclusive.sh
Untracked: code/BothFCnewOInclusive.sh.err
Untracked: code/BothFCnewOInclusive.sh.out
Untracked: code/ChimpStarMM2.err
Untracked: code/ChimpStarMM2.out
Untracked: code/ChimpStarMM2.sh
Untracked: code/ClassifyLeafviz.sh
Untracked: code/ClosestorthoEx.err
Untracked: code/ClosestorthoEx.out
Untracked: code/ClosestorthoEx.sh
Untracked: code/Config_chimp.yaml
Untracked: code/Config_chimp_full.yaml
Untracked: code/Config_human.yaml
Untracked: code/ConvertJunc2Bed.err
Untracked: code/ConvertJunc2Bed.out
Untracked: code/ConvertJunc2Bed.sh
Untracked: code/CountNucleotides.py
Untracked: code/CrossMapChimpRNA.sh
Untracked: code/CrossMapThreeprime.sh
Untracked: code/CrossmapChimp3prime.err
Untracked: code/CrossmapChimp3prime.out
Untracked: code/CrossmapChimpRNA.err
Untracked: code/CrossmapChimpRNA.out
Untracked: code/DTUTR.sh
Untracked: code/DiffDom_RNAmotif_4.err
Untracked: code/DiffDom_RNAmotif_4.out
Untracked: code/DiffDom_RNAmotif_4.sh
Untracked: code/DiffDom_RNAmotif_4_splitDE.err
Untracked: code/DiffDom_RNAmotif_4_splitDE.out
Untracked: code/DiffDom_RNAmotif_4_splitDE.sh
Untracked: code/DiffSplice.err
Untracked: code/DiffSplice.out
Untracked: code/DiffSplice.sh
Untracked: code/DiffSplicePlots.err
Untracked: code/DiffSplicePlots.out
Untracked: code/DiffSplicePlots.sh
Untracked: code/DiffSplicePlots_gencode.sh
Untracked: code/DiffSplice_gencode.sh
Untracked: code/DiffSplice_removebad.err
Untracked: code/DiffSplice_removebad.out
Untracked: code/DiffSplice_removebad.sh
Untracked: code/Filter255.err
Untracked: code/Filter255.out
Untracked: code/Filter255MM.sh
Untracked: code/FilterPrimSec.err
Untracked: code/FilterPrimSec.out
Untracked: code/FilterPrimSec.sh
Untracked: code/FilterReverseLift.err
Untracked: code/FilterReverseLift.out
Untracked: code/FindDomXCutoff.py
Untracked: code/FindIntronForDomPAS.err
Untracked: code/FindIntronForDomPAS.out
Untracked: code/FindIntronForDomPAS.sh
Untracked: code/FindIntronForDomPAS_DF.sh
Untracked: code/GencodeDiffSplice.err
Untracked: code/GencodeDiffSplice.out
Untracked: code/GetMAPQscore.py
Untracked: code/GetSecondaryMap.py
Untracked: code/GetTopminus2Usage.py
Untracked: code/H3K36me3DTplot.err
Untracked: code/H3K36me3DTplot.out
Untracked: code/H3K36me3DTplot.sh
Untracked: code/H3K36me3DTplot_DiffIso.err
Untracked: code/H3K36me3DTplot_DiffIso.out
Untracked: code/H3K36me3DTplot_DiffIso.sh
Untracked: code/H3K36me3DTplot_Specific.err
Untracked: code/H3K36me3DTplot_Specific.out
Untracked: code/H3K36me3DTplot_Specific.sh
Untracked: code/H3K36me3DTplot_distalPAS.err
Untracked: code/H3K36me3DTplot_distalPAS.out
Untracked: code/H3K36me3DTplot_distalPAS.sh
Untracked: code/H3K36me3DTplot_transcript.err
Untracked: code/H3K36me3DTplot_transcript.out
Untracked: code/H3K36me3DTplot_transcript.sh
Untracked: code/H3K36me3DTplotwide.err
Untracked: code/H3K36me3DTplotwide.out
Untracked: code/H3K36me3DTplotwide.sh
Untracked: code/H3K9me3DTplot_transcript.err
Untracked: code/H3K9me3DTplot_transcript.out
Untracked: code/H3K9me3DTplot_transcript.sh
Untracked: code/H3K9me3_processandDT.sh
Untracked: code/HchromOrder.err
Untracked: code/HchromOrder.out
Untracked: code/InfoContentShannon.py
Untracked: code/InfoContentbyInd.py
Untracked: code/IntersectMMandOrtho.err
Untracked: code/IntersectMMandOrtho.out
Untracked: code/IntersectPASandOrtho.err
Untracked: code/IntersectPASandOrtho.out
Untracked: code/JunctionLift.err
Untracked: code/JunctionLift.out
Untracked: code/JunctionLiftFinalChimp.err
Untracked: code/JunctionLiftFinalChimp.out
Untracked: code/Lift5perPAS.sh
Untracked: code/Lift5perPASbed.err
Untracked: code/Lift5perPASbed.out
Untracked: code/LiftClustersFirst.err
Untracked: code/LiftClustersFirst.out
Untracked: code/LiftClustersFirst_remove.err
Untracked: code/LiftClustersFirst_remove.out
Untracked: code/LiftClustersSecond.err
Untracked: code/LiftClustersSecond.out
Untracked: code/LiftClustersSecond_remove.err
Untracked: code/LiftClustersSecond_remove.out
Untracked: code/LiftFinalChimpJunc2Human.sh
Untracked: code/LiftOrthoPAS2chimp.sh
Untracked: code/LiftorthoPAS.err
Untracked: code/LiftorthoPASt.out
Untracked: code/Log.out
Untracked: code/MapBadSamples.err
Untracked: code/MapBadSamples.out
Untracked: code/MapBadSamples.sh
Untracked: code/MapStats.err
Untracked: code/MapStats.out
Untracked: code/MaxEntCode/
Untracked: code/MergeClusters.err
Untracked: code/MergeClusters.out
Untracked: code/MergeClusters.sh
Untracked: code/MismatchNumbers.err
Untracked: code/MismatchNumbers.out
Untracked: code/MismatchNumbers.sh
Untracked: code/NuclearDTUTR.err
Untracked: code/NuclearDTUTRt.out
Untracked: code/NuclearPlotsDEandDiffDom_4.err
Untracked: code/NuclearPlotsDEandDiffDom_4.out
Untracked: code/NuclearPlotsDEandDiffDom_4.sh
Untracked: code/PAS_ATTAAA.err
Untracked: code/PAS_ATTAAA.out
Untracked: code/PAS_ATTAAA.sh
Untracked: code/PAS_ATTAAADF.err
Untracked: code/PAS_ATTAAADF.out
Untracked: code/PAS_ATTAAA_df.sh
Untracked: code/PAS_seqExpanded.sh
Untracked: code/PAS_sequence.err
Untracked: code/PAS_sequence.out
Untracked: code/PAS_sequenceDF.err
Untracked: code/PAS_sequenceDF.out
Untracked: code/PASexpanded_sequenceDF.err
Untracked: code/PASexpanded_sequenceDF.out
Untracked: code/PASsequences.sh
Untracked: code/PASsequences_DF.sh
Untracked: code/PlotNuclearUsagebySpecies.R
Untracked: code/PlotNuclearUsagebySpecies_DF.R
Untracked: code/PlotNuclearUsagebySpecies_DF_4DIC.R
Untracked: code/PlotNuclearUsagebySpecies_DF_DEout.R
Untracked: code/QuantMergeClusters
Untracked: code/QuantMergeClusters.err
Untracked: code/QuantMergeClusters.out
Untracked: code/QuantMergedClusters.sh
Untracked: code/RNATranscriptDTplot.err
Untracked: code/RNATranscriptDTplot.out
Untracked: code/RNATranscriptDTplot.sh
Untracked: code/RNAmotif_PAS.err
Untracked: code/RNAmotif_PAS.out
Untracked: code/RNAmotif_PAS.sh
Untracked: code/RNAmotif_PAS_chimp.err
Untracked: code/RNAmotif_PAS_chimp.out
Untracked: code/RNAmotif_PAS_chimp.sh
Untracked: code/Rev_liftoverPAShg19to38.err
Untracked: code/Rev_liftoverPAShg19to38.out
Untracked: code/ReverseLiftFilter.R
Untracked: code/RunFixCluster.err
Untracked: code/RunFixCluster.out
Untracked: code/RunFixLeafCluster.sh
Untracked: code/RunNegMCMediation.err
Untracked: code/RunNegMCMediation.sh
Untracked: code/RunNegMCMediationDF.err
Untracked: code/RunNegMCMediationDF.out
Untracked: code/RunNegMCMediationDF.sh
Untracked: code/RunNegMCMediationr.out
Untracked: code/RunNewDom.err
Untracked: code/RunNewDom.out
Untracked: code/RunPosMCMediation.err
Untracked: code/RunPosMCMediation.sh
Untracked: code/RunPosMCMediationDF.err
Untracked: code/RunPosMCMediationDF.out
Untracked: code/RunPosMCMediationDF.sh
Untracked: code/RunPosMCMediationr.out
Untracked: code/SAF215upbed_gen.py
Untracked: code/SAF2Bed.py
Untracked: code/Snakefile
Untracked: code/SnakefilePAS
Untracked: code/SnakefilePASfilt
Untracked: code/SortIndexBadSamples.err
Untracked: code/SortIndexBadSamples.out
Untracked: code/SortIndexBadSamples.sh
Untracked: code/StarMM2.err
Untracked: code/StarMM2.out
Untracked: code/StarMM2.sh
Untracked: code/TestFC.err
Untracked: code/TestFC.out
Untracked: code/TestFC.sh
Untracked: code/TotalTranscriptDTplot.err
Untracked: code/TotalTranscriptDTplot.out
Untracked: code/UTR2FASTA.py
Untracked: code/Upstream10Bases_general.py
Untracked: code/allPASSeq_df.sh
Untracked: code/apaQTLsnake.err
Untracked: code/apaQTLsnake.out
Untracked: code/apaQTLsnakePAS.err
Untracked: code/apaQTLsnakePAS.out
Untracked: code/apaQTLsnakePAShuman.err
Untracked: code/apaQTLsnakefiltPAS.err
Untracked: code/apaQTLsnakefiltPAS.out
Untracked: code/assignPeak2Intronicregion.err
Untracked: code/assignPeak2Intronicregion.out
Untracked: code/assignPeak2Intronicregion.sh
Untracked: code/bam2junc.err
Untracked: code/bam2junc.out
Untracked: code/bam2junc_remove.err
Untracked: code/bam2junc_remove.out
Untracked: code/bed215upbed.py
Untracked: code/bed2Bedbothstrand.py
Untracked: code/bed2SAF_gen.py
Untracked: code/bed2saf.py
Untracked: code/bg_to_cov.py
Untracked: code/buildIndecpantro5
Untracked: code/buildIndecpantro5.sh
Untracked: code/buildLeafviz.err
Untracked: code/buildLeafviz.out
Untracked: code/buildLeafviz.sh
Untracked: code/buildLeafviz_leadAnno.sh
Untracked: code/buildLeafviz_leafanno.err
Untracked: code/buildLeafviz_leafanno.out
Untracked: code/buildStarIndex.sh
Untracked: code/callPeaksYL.py
Untracked: code/chimpChromprder.sh
Untracked: code/chimpMultiCov.err
Untracked: code/chimpMultiCov.out
Untracked: code/chimpMultiCov.sh
Untracked: code/chimpMultiCov255.sh
Untracked: code/chimpMultiCovInclusive.err
Untracked: code/chimpMultiCovInclusive.out
Untracked: code/chimpMultiCovInclusive.sh
Untracked: code/chooseAnno2Bed.py
Untracked: code/chooseAnno2SAF.py
Untracked: code/chooseSignalSite.py
Untracked: code/chromOrder.err
Untracked: code/chromOrder.out
Untracked: code/classifyLeafviz.err
Untracked: code/classifyLeafviz.out
Untracked: code/cleanbed2saf.py
Untracked: code/cluster.json
Untracked: code/cluster2bed.py
Untracked: code/clusterLiftReverse.sh
Untracked: code/clusterLiftReverse_removebad.sh
Untracked: code/clusterLiftprimary.sh
Untracked: code/clusterLiftprimary_removebad.sh
Untracked: code/clusterPAS.json
Untracked: code/clusterfiltPAS.json
Untracked: code/comands2Mege.sh
Untracked: code/converBam2Junc.sh
Untracked: code/converBam2Junc_removeBad.sh
Untracked: code/convertNumeric.py
Untracked: code/extraSnakefiltpas
Untracked: code/extractPhaastConGeneral.py
Untracked: code/extractPhyloReg.py
Untracked: code/extractPhyloRegGene.py
Untracked: code/extractPhylopGeneral.py
Untracked: code/extractPhylopGeneral_20way.py
Untracked: code/extractPhylopGeneral_noName.py
Untracked: code/extractPhylopReg200down.py
Untracked: code/extractPhylopReg200up.py
Untracked: code/filter5perc.R
Untracked: code/filter5percPAS.py
Untracked: code/filter5percPheno.py
Untracked: code/filterBamforMP.pysam2_gen.py
Untracked: code/filterJuncChroms.err
Untracked: code/filterJuncChroms.out
Untracked: code/filterMissprimingInNuc10_gen.py
Untracked: code/filterNumChroms.py
Untracked: code/filterPASforMP.py
Untracked: code/filterPostLift.py
Untracked: code/filterPrimaryread.py
Untracked: code/filterSAFforMP_gen.py
Untracked: code/filterSecondaryread.py
Untracked: code/filterSortBedbyCleanedBed_gen.R
Untracked: code/filterpeaks.py
Untracked: code/fixExonFC.py
Untracked: code/fixFChead.py
Untracked: code/fixFChead_bothfrac.py
Untracked: code/fixFCheadforExp.py
Untracked: code/fixLeafCluster.py
Untracked: code/fixLiftedJunc.py
Untracked: code/fixUTRexonanno.py
Untracked: code/formathg38Anno.py
Untracked: code/generateStarIndex.err
Untracked: code/generateStarIndex.out
Untracked: code/generateStarIndexHuman.err
Untracked: code/generateStarIndexHuman.out
Untracked: code/getAlloverlap.py
Untracked: code/getRNAseqMapStats.sh
Untracked: code/hg19MapStats.err
Untracked: code/hg19MapStats.out
Untracked: code/hg19MapStats.sh
Untracked: code/humanChromorder.sh
Untracked: code/humanFiles
Untracked: code/humanMultiCov.err
Untracked: code/humanMultiCov.out
Untracked: code/humanMultiCov.sh
Untracked: code/humanMultiCov255.err
Untracked: code/humanMultiCov255.out
Untracked: code/humanMultiCov255.sh
Untracked: code/humanMultiCovInclusive.err
Untracked: code/humanMultiCovInclusive.out
Untracked: code/humanMultiCov_inclusive.sh
Untracked: code/infoContentSimpson.py
Untracked: code/intersectAnno.err
Untracked: code/intersectAnno.out
Untracked: code/intersectAnnoExt.err
Untracked: code/intersectAnnoExt.out
Untracked: code/intersectLiftedPAS.sh
Untracked: code/leafcutter_merge_regtools_redo.py
Untracked: code/liftJunctionFiles.sh
Untracked: code/liftPAS19to38.sh
Untracked: code/liftVCF.out
Untracked: code/liftVCF.sh
Untracked: code/liftoverPAShg19to38.err
Untracked: code/liftoverPAShg19to38.out
Untracked: code/lliftVCF.err
Untracked: code/log/
Untracked: code/make5percPeakbed.py
Untracked: code/makeDIC.err
Untracked: code/makeDIC.out
Untracked: code/makeFileID.py
Untracked: code/makeNuclearDapaplots.sh
Untracked: code/makeNuclearDapaplots_DF.sh
Untracked: code/makeNuclearPlots.err
Untracked: code/makeNuclearPlots.out
Untracked: code/makeNuclearPlotsDF.err
Untracked: code/makeNuclearPlotsDF.out
Untracked: code/makePheno.py
Untracked: code/makeSamplyGroupsChimp_TvN.py
Untracked: code/makeSamplyGroupsHuman_TvN.py
Untracked: code/makedICPlots_DF.sh
Untracked: code/mapRNAseqhg19.sh
Untracked: code/mapRNAseqhg19_newPipeline.sh
Untracked: code/maphg19.err
Untracked: code/maphg19.out
Untracked: code/maphg19.sh
Untracked: code/maphg19_new.err
Untracked: code/maphg19_new.out
Untracked: code/maphg19_sub.err
Untracked: code/maphg19_sub.out
Untracked: code/maphg19_subjunc.sh
Untracked: code/mediation_test.R
Untracked: code/merge.err
Untracked: code/mergeChimp3prime_inhg38.sh
Untracked: code/mergeChimpRNA.sh
Untracked: code/merge_leafcutter_clusters_redo.py
Untracked: code/mergeandBWRNAseq.sh
Untracked: code/mergeandsort_ChimpinHuman.err
Untracked: code/mergeandsort_ChimpinHuman.out
Untracked: code/mergeandsort_H3K9me3
Untracked: code/mergeandsort_h3k36me3
Untracked: code/mergeandsorth3k36me3.sh
Untracked: code/mergedBam2BW.sh
Untracked: code/mergedbam2bw.err
Untracked: code/mergedbam2bw.out
Untracked: code/mergedbamRNAand2bw.err
Untracked: code/mergedbamRNAand2bw.out
Untracked: code/nameClusters.py
Untracked: code/namePeaks.py
Untracked: code/negativeMediation_montecarlo.R
Untracked: code/negativeMediation_montecarloDF.R
Untracked: code/nuclearTranscriptDTplot.err
Untracked: code/nuclearTranscriptDTplot.out
Untracked: code/numMultimap.py
Untracked: code/overlapMMandOrthoexon.sh
Untracked: code/overlapPAS.err
Untracked: code/overlapPAS.out
Untracked: code/overlapPASandOrthoexon.sh
Untracked: code/overlapapaQTLPAS.sh
Untracked: code/overlapapaQTLPAS_extended.sh
Untracked: code/overlapapaQTLPAS_samples.sh
Untracked: code/parseHg38.py
Untracked: code/peak2PAS.py
Untracked: code/pheno2countonly.R
Untracked: code/postiveMediation_montecarlo.R
Untracked: code/postiveMediation_montecarlo_DF.R
Untracked: code/prepareAnnoLeafviz.err
Untracked: code/prepareAnnoLeafviz.out
Untracked: code/prepareCleanLiftedFC_5perc4LC.py
Untracked: code/prepareLeafvizAnno.sh
Untracked: code/preparePAS4lift.py
Untracked: code/prepare_phenotype_table.py
Untracked: code/primaryLift.err
Untracked: code/primaryLift.out
Untracked: code/primaryLift.sh
Untracked: code/processhg38exons.py
Untracked: code/quantJunc.sh
Untracked: code/quantJunc_TEST.sh
Untracked: code/quantJunc_removeBad.sh
Untracked: code/quantLiftedPAS.err
Untracked: code/quantLiftedPAS.out
Untracked: code/quantLiftedPAS.sh
Untracked: code/quantLiftedPASPrimary.err
Untracked: code/quantLiftedPASPrimary.out
Untracked: code/quantLiftedPASPrimary.sh
Untracked: code/quatJunc.err
Untracked: code/quatJunc.out
Untracked: code/recChimpback2Human.err
Untracked: code/recChimpback2Human.out
Untracked: code/recLiftchim2human.sh
Untracked: code/revLift.err
Untracked: code/revLift.out
Untracked: code/revLiftPAShg38to19.sh
Untracked: code/reverseLift.sh
Untracked: code/runCheckReverseLift.sh
Untracked: code/runChimpDiffIso.sh
Untracked: code/runChimpDiffIsoDF.sh
Untracked: code/runCountNucleotides.err
Untracked: code/runCountNucleotides.out
Untracked: code/runCountNucleotides.sh
Untracked: code/runCountNucleotidesPantro6.err
Untracked: code/runCountNucleotidesPantro6.out
Untracked: code/runCountNucleotides_pantro6.sh
Untracked: code/runFilterNumChroms.sh
Untracked: code/runHumanDiffIso.sh
Untracked: code/runHumanDiffIsoDF.sh
Untracked: code/runNewDom.sh
Untracked: code/runNuclearDiffIso_DF.sh
Untracked: code/runNuclearDifffIso.sh
Untracked: code/runTotalDiffIso.sh
Untracked: code/run_Chimpleafcutter_ds.err
Untracked: code/run_Chimpleafcutter_ds.out
Untracked: code/run_Chimpverifybam.err
Untracked: code/run_Chimpverifybam.out
Untracked: code/run_Humanleafcutter_dF.err
Untracked: code/run_Humanleafcutter_dF.out
Untracked: code/run_Humanleafcutter_ds.err
Untracked: code/run_Humanleafcutter_ds.out
Untracked: code/run_Nuclearleafcutter_ds.err
Untracked: code/run_Nuclearleafcutter_ds.out
Untracked: code/run_Nuclearleafcutter_dsDF.err
Untracked: code/run_Nuclearleafcutter_dsDF.out
Untracked: code/run_Totalleafcutter_ds.err
Untracked: code/run_Totalleafcutter_ds.out
Untracked: code/run_chimpverifybam.sh
Untracked: code/run_verifyBam.sh
Untracked: code/run_verifybam.err
Untracked: code/run_verifybam.out
Untracked: code/slurm-62824013.out
Untracked: code/slurm-62825841.out
Untracked: code/slurm-62826116.out
Untracked: code/slurm-64108209.out
Untracked: code/slurm-64108521.out
Untracked: code/slurm-64108557.out
Untracked: code/snakePASChimp.err
Untracked: code/snakePASChimp.out
Untracked: code/snakePAShuman.out
Untracked: code/snakemake.batch
Untracked: code/snakemakeChimp.err
Untracked: code/snakemakeChimp.out
Untracked: code/snakemakeHuman.err
Untracked: code/snakemakeHuman.out
Untracked: code/snakemakePAS.batch
Untracked: code/snakemakePASFiltChimp.err
Untracked: code/snakemakePASFiltChimp.out
Untracked: code/snakemakePASFiltHuman.err
Untracked: code/snakemakePASFiltHuman.out
Untracked: code/snakemakePAS_Human.batch
Untracked: code/snakemakePASchimp.batch
Untracked: code/snakemakePAShuman.batch
Untracked: code/snakemake_chimp.batch
Untracked: code/snakemake_human.batch
Untracked: code/snakemakefiltPAS.batch
Untracked: code/snakemakefiltPAS_chimp.sh
Untracked: code/snakemakefiltPAS_human.batch
Untracked: code/snakemakefiltPAS_human.sh
Untracked: code/spliceSite2Fasta.py
Untracked: code/submit-snakemake-chimp.sh
Untracked: code/submit-snakemake-human.sh
Untracked: code/submit-snakemakePAS-chimp.sh
Untracked: code/submit-snakemakePAS-human.sh
Untracked: code/submit-snakemakefiltPAS-chimp.sh
Untracked: code/submit-snakemakefiltPAS-human.sh
Untracked: code/subsetChr.py
Untracked: code/subset_diffisopheno.py
Untracked: code/subset_diffisopheno_Chimp_tvN.py
Untracked: code/subset_diffisopheno_Chimp_tvN_DF.py
Untracked: code/subset_diffisopheno_Huma_tvN.py
Untracked: code/subset_diffisopheno_Huma_tvN_DF.py
Untracked: code/subset_diffisopheno_Nuclear_HvC.py
Untracked: code/subset_diffisopheno_Nuclear_HvC_DF.py
Untracked: code/subset_diffisopheno_Total_HvC.py
Untracked: code/test
Untracked: code/test.txt
Untracked: code/threeprimeOrthoFC.out
Untracked: code/threeprimeOrthoFC.sh
Untracked: code/threeprimeOrthoFCcd.err
Untracked: code/transcriptDTplotsNuclear.sh
Untracked: code/transcriptDTplotsTotal.sh
Untracked: code/tripseq-analysis/
Untracked: code/verifyBam4973.sh
Untracked: code/verifyBam4973inHuman.sh
Untracked: code/verifybam4973.err
Untracked: code/verifybam4973.out
Untracked: code/verifybam4973HumanMap.err
Untracked: code/verifybam4973HumanMap.out
Untracked: code/wrap_Chimpverifybam.err
Untracked: code/wrap_Chimpverifybam.out
Untracked: code/wrap_chimpverifybam.sh
Untracked: code/wrap_verifyBam.sh
Untracked: code/wrap_verifybam.err
Untracked: code/wrap_verifybam.out
Untracked: code/writeMergecode.py
Untracked: data/._.DS_Store
Untracked: data/._HC_filenames.txt
Untracked: data/._HC_filenames.txt.sb-4426323c-IKIs0S
Untracked: data/._HC_filenames.xlsx
Untracked: data/._MapPantro6_meta.txt
Untracked: data/._MapPantro6_meta.txt.sb-a5794dd2-Cskmlm
Untracked: data/._MapPantro6_meta.xlsx
Untracked: data/._OppositeSpeciesMap.txt
Untracked: data/._OppositeSpeciesMap.txt.sb-a5794dd2-mayWJf
Untracked: data/._OppositeSpeciesMap.xlsx
Untracked: data/._RNASEQ_metadata.txt
Untracked: data/._RNASEQ_metadata.txt.sb-4426323c-TE4ns3
Untracked: data/._RNASEQ_metadata.txt.sb-51f67ae1-HXp7Gq
Untracked: data/._RNASEQ_metadata_2Removed.txt
Untracked: data/._RNASEQ_metadata_2Removed.txt.sb-4426323c-a4lBwx
Untracked: data/._RNASEQ_metadata_2Removed.xlsx
Untracked: data/._RNASEQ_metadata_stranded.txt
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-a5794dd2-D659m2
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-a5794dd2-ImNMoY
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-e4bf31f0-ZGnGgl
Untracked: data/._RNASEQ_metadata_stranded.xlsx
Untracked: data/._TrialFiltersMeta.txt
Untracked: data/._TrialFiltersMeta.txt.sb-9845453e-R58Y0Q
Untracked: data/._metadata_HCpanel.txt
Untracked: data/._metadata_HCpanel.txt.sb-a3d92a2d-b9cYoF
Untracked: data/._metadata_HCpanel.txt.sb-a5794dd2-i594qs
Untracked: data/._metadata_HCpanel.txt.sb-f4823d1e-qihGek
Untracked: data/._metadata_HCpanel_frompantro5.xlsx
Untracked: data/._~$RNASEQ_metadata.xlsx
Untracked: data/._~$metadata_HCpanel.xlsx
Untracked: data/._.xlsx
Untracked: data/ALU/
Untracked: data/AREelements/
Untracked: data/BaseComp/
Untracked: data/CleanLiftedPeaks_FC_primary/
Untracked: data/CompapaQTLpas/
Untracked: data/DIC_Viz/
Untracked: data/DNDS/
Untracked: data/DTmatrix/
Untracked: data/DiffDomandDE_example/
Untracked: data/DiffExpression/
Untracked: data/DiffIso_Nuclear/
Untracked: data/DiffIso_Nuclear_DF/
Untracked: data/DiffIso_Total/
Untracked: data/DiffSplice/
Untracked: data/DiffSplice_liftedJunc/
Untracked: data/DiffSplice_removeBad/
Untracked: data/DifferentialPASUsageResults.txt
Untracked: data/DistTwoDom/
Untracked: data/DomDefGreaterX/
Untracked: data/DomStructure_4/
Untracked: data/DominantPAS/
Untracked: data/DominantPAS_DF/
Untracked: data/DoubleFilterUsageNumeric/
Untracked: data/EvalPantro5/
Untracked: data/H3K36me3/
Untracked: data/HC_filenames.txt
Untracked: data/HC_filenames.xlsx
Untracked: data/HumanMolPheno/
Untracked: data/HumanUTR/
Untracked: data/IndInfoContent/
Untracked: data/InfoContent/
Untracked: data/Khan_prot/
Untracked: data/Li_eqtls/
Untracked: data/MapPantro6_meta.txt
Untracked: data/MapPantro6_meta.xlsx
Untracked: data/MapStats/
Untracked: data/NormalizedClusters/
Untracked: data/NuclearHvC/
Untracked: data/NuclearHvC_DF/
Untracked: data/OppositeSpeciesMap.txt
Untracked: data/OppositeSpeciesMap.xlsx
Untracked: data/OrthoExonBed/
Untracked: data/OverlapBenchmark/
Untracked: data/OverlappingPAS/
Untracked: data/PAS/
Untracked: data/PAS_SAF/
Untracked: data/PAS_doubleFilter/
Untracked: data/PTM/
Untracked: data/Peaks_5perc/
Untracked: data/PhastCon/
Untracked: data/Pheno_5perc/
Untracked: data/Pheno_5perc_DF_nuclear/
Untracked: data/Pheno_5perc_nuclear/
Untracked: data/Pheno_5perc_nuclear_old/
Untracked: data/Pheno_5perc_total/
Untracked: data/PhyloP/
Untracked: data/Pol2Chip/
Untracked: data/QTLPASoverlap/
Untracked: data/RNASEQ_metadata.txt
Untracked: data/RNASEQ_metadata_2Removed.txt
Untracked: data/RNASEQ_metadata_2Removed.xlsx
Untracked: data/RNASEQ_metadata_stranded.txt
Untracked: data/RNASEQ_metadata_stranded.txt.sb-e4bf31f0-ZGnGgl/
Untracked: data/RNASEQ_metadata_stranded.xlsx
Untracked: data/RegElements/
Untracked: data/SignalSites/
Untracked: data/SignalSites_doublefilter/
Untracked: data/SpliceSite/
Untracked: data/TestAnnoBiasOE/
Untracked: data/TestMM2/
Untracked: data/TestMM2_AS/
Untracked: data/TestMM2_PrimaryRead/
Untracked: data/TestMM2_SeondaryRead/
Untracked: data/TestMM2_mismatch/
Untracked: data/TestMM2_quality/
Untracked: data/TestWithinMergePAS/
Untracked: data/Test_FC_methods/
Untracked: data/TestedNuclearapaQTLGenes.txt
Untracked: data/Threeprime2Ortho/
Untracked: data/TotalFractionPAS/
Untracked: data/TotalHvC/
Untracked: data/TrialFiltersMeta.txt
Untracked: data/TwoBadSampleAnalysis/
Untracked: data/UnliftedSites/
Untracked: data/UrichElements/
Untracked: data/Wang_ribo/
Untracked: data/apaQTLGenes/
Untracked: data/bioGRID/
Untracked: data/chainFiles/
Untracked: data/cleanPeaks_anno/
Untracked: data/cleanPeaks_byspecies/
Untracked: data/cleanPeaks_lifted/
Untracked: data/files4viz_nuclear/
Untracked: data/files4viz_nuclear_DF/
Untracked: data/greatapa/
Untracked: data/gsea/
Untracked: data/gviz/
Untracked: data/hypoxia/
Untracked: data/leafviz/
Untracked: data/liftover_files/
Untracked: data/mediation/
Untracked: data/mediation_DF/
Untracked: data/metadata_HCpanel.txt
Untracked: data/metadata_HCpanel.xlsx
Untracked: data/metadata_HCpanel_extra.txt
Untracked: data/metadata_HCpanel_frompantro5.txt
Untracked: data/metadata_HCpanel_frompantro5.xlsx
Untracked: data/miRNA/
Untracked: data/multimap/
Untracked: data/orthoUTR/
Untracked: data/paiDecay/
Untracked: data/primaryLift/
Untracked: data/randomIntronCons/
Untracked: data/reverseLift/
Untracked: data/testQuant/
Untracked: data/utrDB/
Untracked: data/~$RNASEQ_metadata.xlsx
Untracked: data/~$metadata_HCpanel.xlsx
Untracked: data/.xlsx
Untracked: output/._.DS_Store
Untracked: output/AUcount_density.pdf
Untracked: output/AUcount_density_sm.pdf
Untracked: output/DEandAPA.txt
Untracked: output/DEandAPA_sig.txt
Untracked: output/DEandTEeffectsize
Untracked: output/DEandTEeffectsize.pdf
Untracked: output/DEeffectsize
Untracked: output/DEeffectsize.pdf
Untracked: output/FigureDF/
Untracked: output/FigurePresColors/
Untracked: output/PropSamesdom
Untracked: output/PropSamesdom.pdf
Untracked: output/SupplementaryTable/
Untracked: output/TEeffectsize
Untracked: output/TEeffectsize.pdf
Untracked: output/Total_DEeffectsize
Untracked: output/Total_DEeffectsize.pdf
Untracked: output/Total_DEeffectsizeNotJusttop.pdf
Untracked: output/Total_DErelationship.pdf
Untracked: output/Total_TEeffectsize
Untracked: output/Total_TEeffectsize.pdf
Untracked: output/Ubiqplot
Untracked: output/Ubiqplot.pdf
Untracked: output/dAPAandDomEnrich.png
Untracked: output/dEandDomEnrich.png
Untracked: output/dediffdom.pdf
Untracked: output/dpnotDE
Untracked: output/dtPlots/
Untracked: output/exandte
Untracked: output/fig1.pdf
Untracked: output/fig2.pdf
Untracked: output/fig3.pdf
Untracked: output/fig4.pdf
Untracked: output/fig5.pdf
Untracked: output/fig6.pdf
Untracked: output/piecharts
Untracked: output/piecharts.pdf
Untracked: output/removeUnilift_fig6 copy.pdf
Untracked: output/removeUnilift_fig6.pdf
Untracked: output/removeUnlift_fig3.pdf
Untracked: output/removeUnlift_fig4.pdf
Untracked: output/removeUnlift_fig5.pdf
Untracked: output/revisions/
Untracked: output/simpson.pdf
Untracked: output/simpson_alTogether.pdf
Untracked: output/supplement/
Untracked: output/whichSiteplot.pdf
Untracked: projectNotes.Rmd
Untracked: proteinModelSet.Rmd
Unstaged changes:
Modified: analysis/ConsPhlop20.Rmd
Modified: analysis/DeandNumPAS.Rmd
Modified: analysis/DirSelectionKhan.Rmd
Modified: analysis/ExploredAPA.Rmd
Modified: analysis/ExploredAPA_DF.Rmd
Modified: analysis/MMExpreiment.Rmd
Modified: analysis/OppositeMap.Rmd
Modified: analysis/PTM_analysis.Rmd
Modified: analysis/ResultsNoUnlifted.Rmd
Modified: analysis/SetsdAPADIC.Rmd
Modified: analysis/TotalDomStructure.Rmd
Modified: analysis/TotalVNuclearBothSpecies.Rmd
Modified: analysis/additionalGSEA.Rmd
Modified: analysis/annotationInfo.Rmd
Modified: analysis/changeMisprimcut.Rmd
Modified: analysis/comp2apaQTLPAS.Rmd
Modified: analysis/correlationPhenos.Rmd
Modified: analysis/dInforContent.Rmd
Modified: analysis/df_QC.Rmd
Modified: analysis/diffExpression.Rmd
Modified: analysis/establishCutoffs.Rmd
Modified: analysis/incorporateQTLsAncestral.Rmd
Modified: analysis/index.Rmd
Modified: analysis/infoContent.Rmd
Modified: analysis/investigatePantro5.Rmd
Modified: analysis/mRNADecay.Rmd
Modified: analysis/miRNAanalysis.Rmd
Modified: analysis/multiMap.Rmd
Modified: analysis/phastCon.Rmd
Modified: analysis/pol2.Rmd
Modified: analysis/speciesSpecific.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/phylopRegElements.Rmd) and HTML (docs/phylopRegElements.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | b769a17 | brimittleman | 2021-01-18 | add 200 up analysis |
| html | f2e7ead | brimittleman | 2020-12-24 | Build site. |
| Rmd | a8cf928 | brimittleman | 2020-12-23 | add sig tests |
| html | d5febd5 | brimittleman | 2020-12-21 | Build site. |
| Rmd | d1e428d | brimittleman | 2020-12-21 | add intron and exon |
| html | 0f74991 | brimittleman | 2020-12-21 | Build site. |
| Rmd | 2cfbc6d | brimittleman | 2020-12-21 | add phylop reg element analysis |
library(workflowr)This is workflowr version 1.6.2
Run ?workflowr for help getting startedlibrary()
library(cowplot)
********************************************************Note: As of version 1.0.0, cowplot does not change the default ggplot2 theme anymore. To recover the previous behavior, execute:
theme_set(theme_cowplot())********************************************************library(tidyverse)── Attaching packages ────────────────────────────────── tidyverse 1.3.0 ──✓ ggplot2 3.2.1 ✓ purrr 0.3.4
✓ tibble 2.1.3 ✓ dplyr 0.8.3
✓ tidyr 1.1.0 ✓ stringr 1.4.0
✓ readr 1.3.1 ✓ forcats 0.4.0── Conflicts ───────────────────────────────────── tidyverse_conflicts() ──
x dplyr::filter() masks stats::filter()
x dplyr::lag() masks stats::lag()I will download regulatory elements and compare them to the PAS regions. I will run this for phylop20 and phlylop100
/project2/gilad/briana/Comparative_APA/data/RegElements
Download from table browser:
- refseq elements
- sno and miRNAs
python extractPhylopGeneral_20way.py ../data/RegElements/hg38_RefSeqElements.bed ../data/RegElements/hg38_RefSeqElements.phyloP20.txt
python extractPhylopGeneral_20way.py ../data/RegElements/hg38_snomiRNA.bed ../data/RegElements/hg38_snomiRNAphyloP20.txt
python extractPhylopGeneral.py ../data/RegElements/hg38_RefSeqElements.bed ../data/RegElements/hg38_RefSeqElements.phyloP100.txt
python extractPhylopGeneral.py ../data/RegElements/hg38_snomiRNA.bed ../data/RegElements/hg38_snomiRNAphyloP100.txt
PAS_20=read.table("../data/PhyloP/PAS_Phylop20.txt", col.names = c("chr","start","end","PAS", "phyloP20"), stringsAsFactors = F) %>% drop_na() %>% mutate(element="PAS") %>% select(element,phyloP20)
PAS_100=read.table("../data/PhyloP/PAS_phyloP.txt", col.names = c("chr","start","end", "phyloP100"), stringsAsFactors = F) %>% drop_na() %>% mutate(element="PAS") %>% select(element,phyloP100)Reg elements:
Reg_20= read.table("../data/RegElements/hg38_RefSeqElements.phyloP20.txt", col.names = c('chr', 'start', 'end', 'element', 'phyloP20' ), header = F, stringsAsFactors = F) %>% drop_na()%>% select(element, phyloP20)
RNAs_20=read.table("../data/RegElements/hg38_snomiRNAphyloP20.txt", col.names = c('chr', 'start', 'end', 'elementName', 'phyloP20' ), header = F, stringsAsFactors = F)%>% mutate(element="smallRNA") %>% drop_na()%>% select(element, phyloP20)
Reg_100= read.table("../data/RegElements/hg38_RefSeqElements.phyloP100.txt", col.names = c('chr', 'start', 'end', 'element', 'phyloP100' ), header = F, stringsAsFactors = F) %>% drop_na()%>% select(element, phyloP100)
RNAs_100=read.table("../data/RegElements/hg38_snomiRNAphyloP100.txt", col.names = c('chr', 'start', 'end', 'element_name', 'phyloP100' ), header = F, stringsAsFactors = F) %>% mutate(element="smallRNA") %>% drop_na() %>% select(element, phyloP100)Look at the elements in the reg and decide which to include:
elementcount=Reg_20 %>% group_by(element) %>% summarise(nele=n()) %>% arrange(desc(nele))
regs_use=c("conserved_region", "enhancer", "CpG_island", "promoter", "silencer", "repeat_instability_region")Will include: conserved_region, enhancer,CpG_island, promoter, silencer, repeat_instability_region
Reg_20_sm=Reg_20 %>% filter(element %in% regs_use)
Reg_100_sm=Reg_100 %>% filter(element %in% regs_use)
RegandRNA_20=Reg_20_sm %>% bind_rows(RNAs_20) %>% bind_rows(PAS_20) %>% mutate(fillcol=ifelse(element=="PAS", "yes", "no"))
RegandRNA_100=Reg_100_sm %>% bind_rows(RNAs_100) %>% bind_rows(PAS_100)%>% mutate(fillcol=ifelse(element=="PAS", "yes", "no"))Plot:
phylop20plot=ggplot(RegandRNA_20, aes(x=element, y=phyloP20,fill=fillcol)) + geom_boxplot() + theme_classic() + labs(title="Regulatory element conservation", y="PhyloP 20 ways", x="")+ theme(axis.text.x = element_text(angle = 90), legend.position ="none")+ scale_fill_brewer(palette = "RdYlBu")
phylop20plot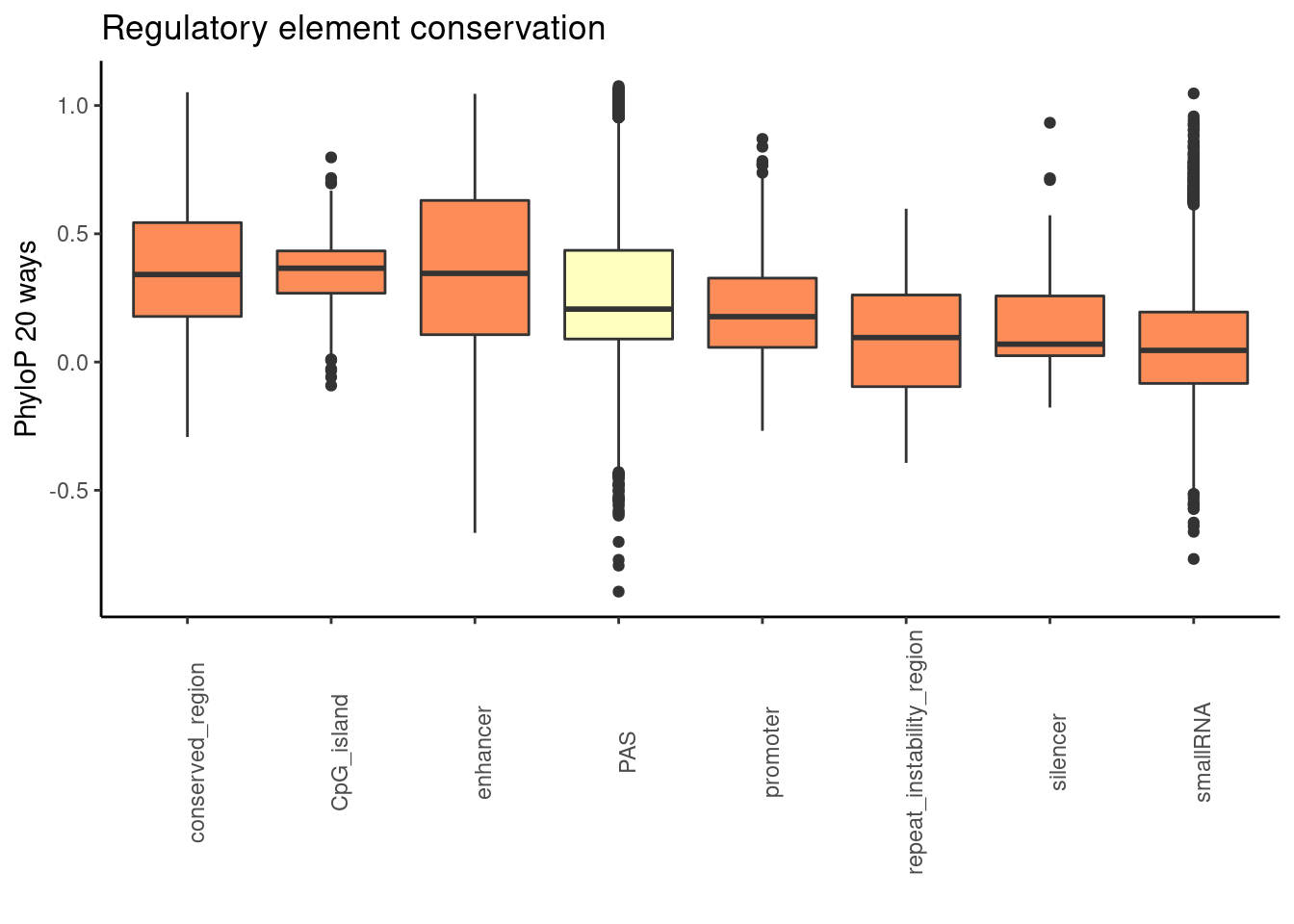
| Version | Author | Date |
|---|---|---|
| 0f74991 | brimittleman | 2020-12-21 |
phylop100plot= ggplot(RegandRNA_100, aes(x=element, y=phyloP100,fill=fillcol)) + geom_boxplot() + theme_classic() + labs(title="Regulatory element conservation", y="PhyloP 100 ways", x="")+ theme(axis.text.x = element_text(angle = 90), legend.position ="none")+ scale_fill_brewer(palette = "RdYlBu")
phylop100plot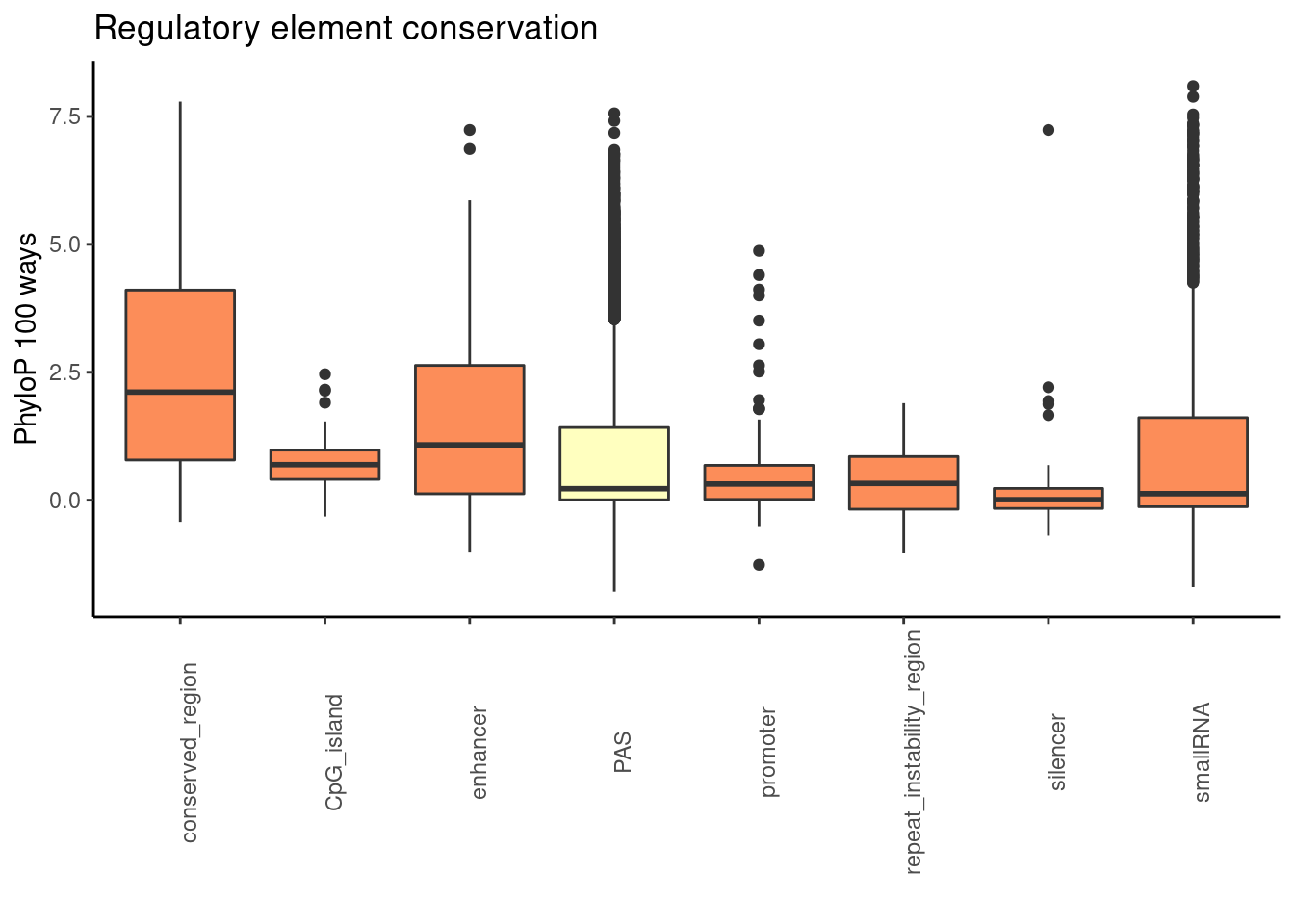
| Version | Author | Date |
|---|---|---|
| 0f74991 | brimittleman | 2020-12-21 |
both_together=plot_grid(phylop20plot,phylop100plot, nrow = 2, labels=c("A", "B"))
both_together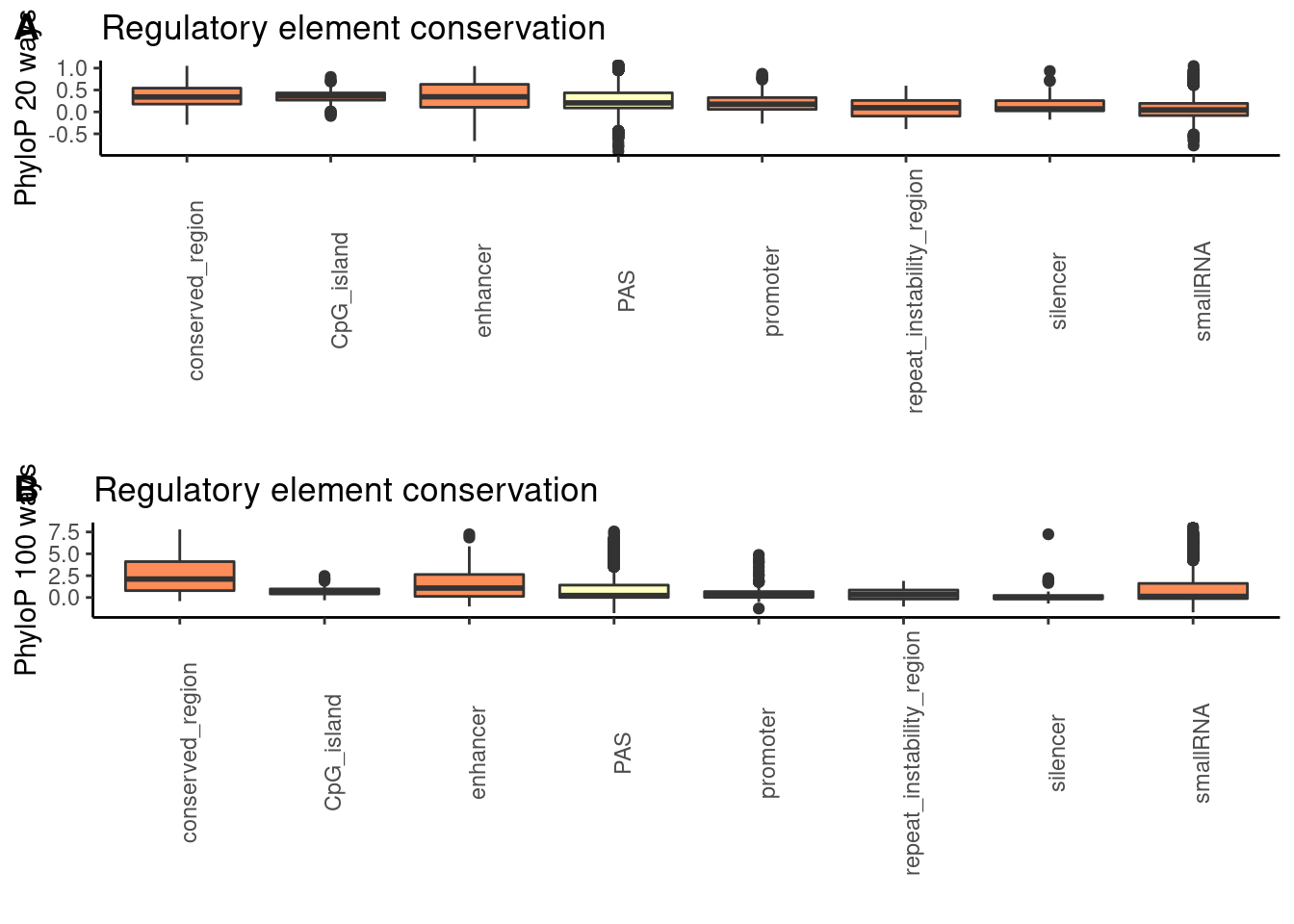
pdf("../output/supplement/Revision-phylopRed.pdf", height=8, width=6,useKerning=F)
both_together
dev.off()png
2 Add introns and exon:
get chr 1-22 from intron and exon file
subsetChr.py
main(fin, fout):
small=open(fout, "w")
chrkeep=["chr1", "chr2", 'chr3', 'chr4', 'chr5', 'chr6', 'chr7', 'chr8', 'chr9', 'chr10', 'chr11', 'chr12', 'chr13', 'chr14', 'chr15', 'chr16', 'chr17', 'chr18', 'chr19', 'chr20', 'chr21','chr22']
for ln in open(fin,"r"):
chrom=ln.split()[0]
if chrom in chrkeep:
small.write(ln)
small.close()
if __name__ == "__main__":
import sys
infile = sys.argv[1]
Outfile =sys.argv[2]
main(infile, Outfile)python subsetChr.py ../data/RegElements/hg38_ncbiRefseq_Formatted_Allannotation_noSNO.Resort.bed ../data/RegElements/hg38_ncbiRefseq_Formatted_Allannotation_noSNO.Resort.1.22.bed
python extractPhylopGeneral_20way.py ../data/RegElements/hg38_ncbiRefseq_Formatted_Allannotation_noSNO.Resort.1.22.bed ../data/RegElements/phylop20_ncbi.txt
Allloc_20=read.table("../data/RegElements/phylop20_ncbi.txt", stringsAsFactors = F, header = F, col.names = c("chr", "start","end", "name", "phyloP20")) %>% separate(name, into=c("element", "gene"),sep=":") %>% drop_na()
ggplot(Allloc_20, aes(x=element, y=phyloP20)) + geom_boxplot() + theme_classic()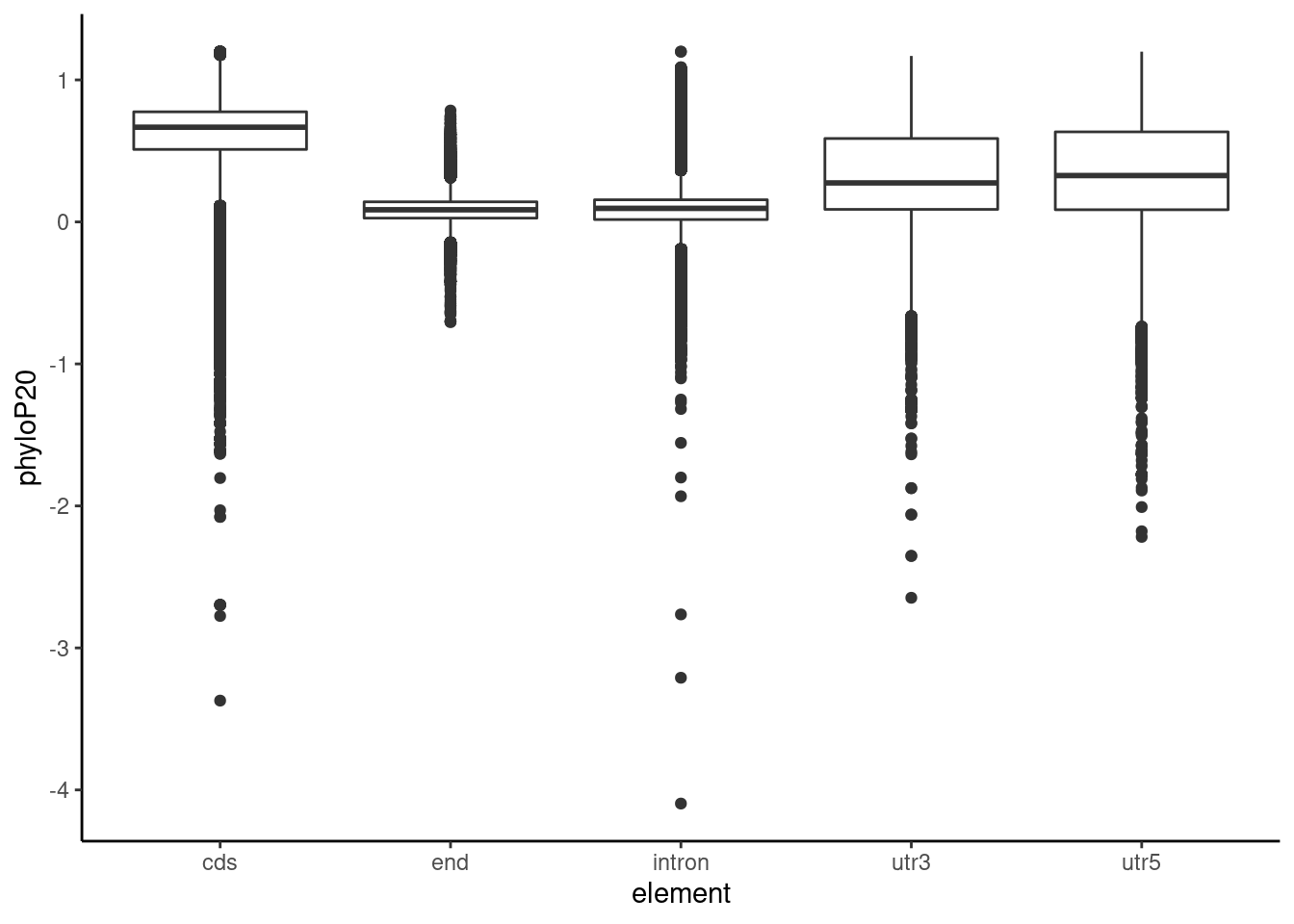
Intron_20=Allloc_20 %>% filter(element=="intron") %>% select(element, phyloP20)
RegandRNAintron_20=Reg_20_sm %>% bind_rows(RNAs_20) %>% bind_rows(PAS_20) %>% bind_rows(Intron_20) %>% mutate(fillcol=ifelse(element=="PAS", "yes", "no"))
ggplot(RegandRNAintron_20, aes(x=element, y=phyloP20,fill=fillcol)) + geom_boxplot() + theme_classic() + labs(title="Regulatory element conservation", y="PhyloP 20 ways", x="")+ theme(axis.text.x = element_text(angle = 90), legend.position ="none")+ scale_fill_brewer(palette = "RdYlBu")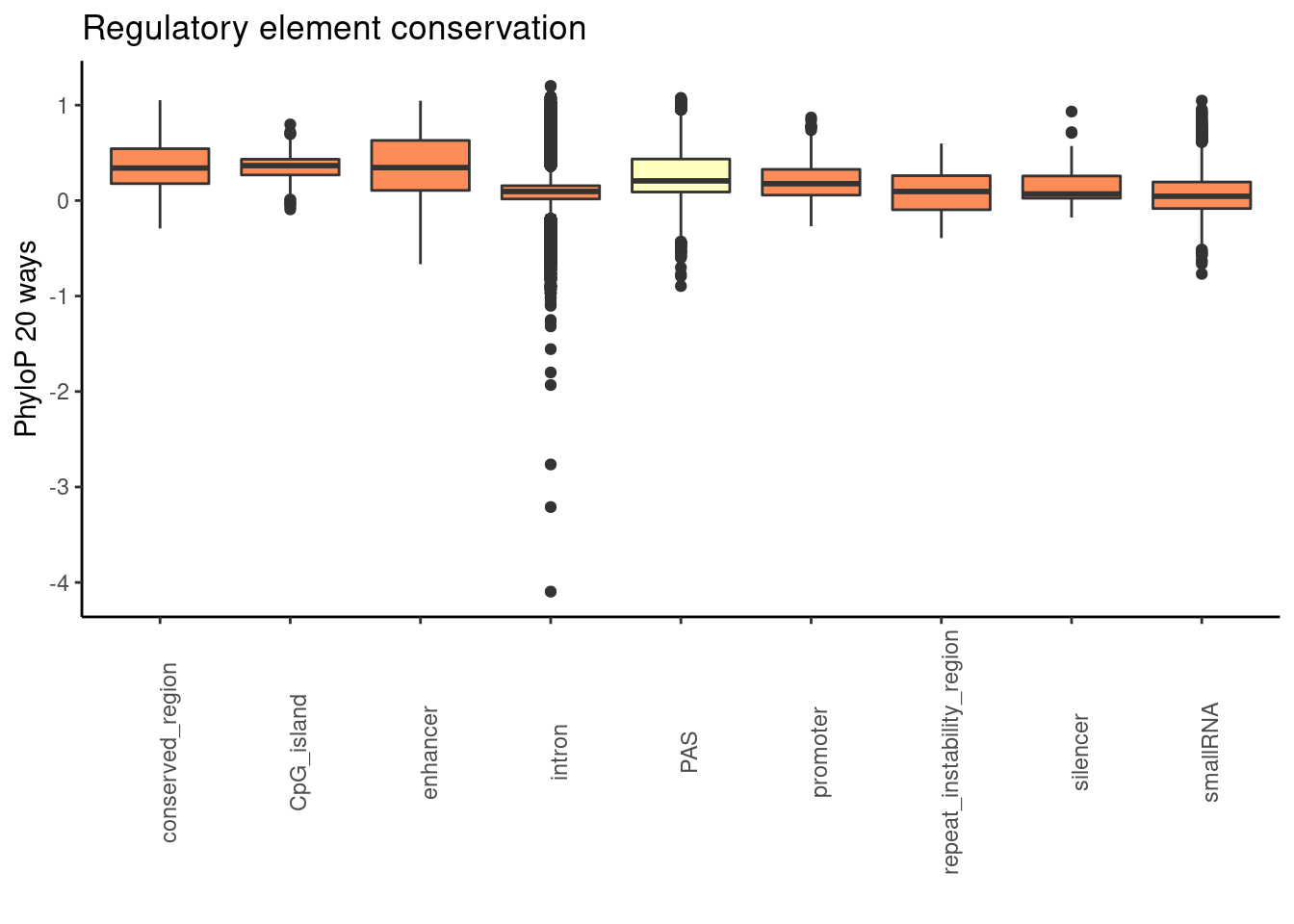
| Version | Author | Date |
|---|---|---|
| f2e7ead | brimittleman | 2020-12-24 |
Significance:
promoter20= RegandRNA_20 %>% filter(element=="promoter")
enhan20= RegandRNA_20 %>% filter(element=="enhancer")
consv20= RegandRNA_20 %>% filter(element=="conserved_region")
cpg20= RegandRNA_20 %>% filter(element=="CpG_island")
repeate20= RegandRNA_20 %>% filter(element=="repeat_instability_region")
silencer20= RegandRNA_20 %>% filter(element=="silencer")
smallRNA20= RegandRNA_20 %>% filter(element=="smallRNA")
wilcox.test(PAS_20$phyloP20,promoter20$phyloP20, alternative = "greater")
Wilcoxon rank sum test with continuity correction
data: PAS_20$phyloP20 and promoter20$phyloP20
W = 5925035, p-value = 0.00064
alternative hypothesis: true location shift is greater than 0wilcox.test(PAS_20$phyloP20,enhan20$phyloP20, alternative = "greater")
Wilcoxon rank sum test with continuity correction
data: PAS_20$phyloP20 and enhan20$phyloP20
W = 26069436, p-value = 1
alternative hypothesis: true location shift is greater than 0wilcox.test(PAS_20$phyloP20,consv20$phyloP20, alternative = "greater")
Wilcoxon rank sum test with continuity correction
data: PAS_20$phyloP20 and consv20$phyloP20
W = 2078060, p-value = 1
alternative hypothesis: true location shift is greater than 0wilcox.test(PAS_20$phyloP20,cpg20$phyloP20, alternative = "greater")
Wilcoxon rank sum test with continuity correction
data: PAS_20$phyloP20 and cpg20$phyloP20
W = 1952657, p-value = 1
alternative hypothesis: true location shift is greater than 0wilcox.test(PAS_20$phyloP20,repeate20$phyloP20, alternative = "greater")
Wilcoxon rank sum test with continuity correction
data: PAS_20$phyloP20 and repeate20$phyloP20
W = 1352876, p-value = 2.014e-05
alternative hypothesis: true location shift is greater than 0wilcox.test(PAS_20$phyloP20,silencer20$phyloP20, alternative = "greater")
Wilcoxon rank sum test with continuity correction
data: PAS_20$phyloP20 and silencer20$phyloP20
W = 1374908, p-value = 0.0002568
alternative hypothesis: true location shift is greater than 0wilcox.test(PAS_20$phyloP20,smallRNA20$phyloP20, alternative = "greater")
Wilcoxon rank sum test with continuity correction
data: PAS_20$phyloP20 and smallRNA20$phyloP20
W = 250749904, p-value < 2.2e-16
alternative hypothesis: true location shift is greater than 0Allloc_100=read.table("../data/RegElements/phylop100_ncbi.txt", stringsAsFactors = F, header = F, col.names = c("chr", "start","end", "name", "phyloP100")) %>% separate(name, into=c("element", "gene"),sep=":") %>% drop_na()
Intron_100=Allloc_100 %>% filter(element=="intron") %>% select(element, phyloP100)
RegandRNAintron_100=Reg_100_sm %>% bind_rows(RNAs_100) %>% bind_rows(PAS_100) %>% bind_rows(Intron_100) %>% mutate(fillcol=ifelse(element=="PAS", "yes", "no"))
ggplot(RegandRNAintron_100, aes(x=element, y=phyloP100,fill=fillcol)) + geom_boxplot() + theme_classic() + labs(title="Regulatory element conservation", y="PhyloP 100 ways", x="")+ theme(axis.text.x = element_text(angle = 90), legend.position ="none")+ scale_fill_brewer(palette = "RdYlBu")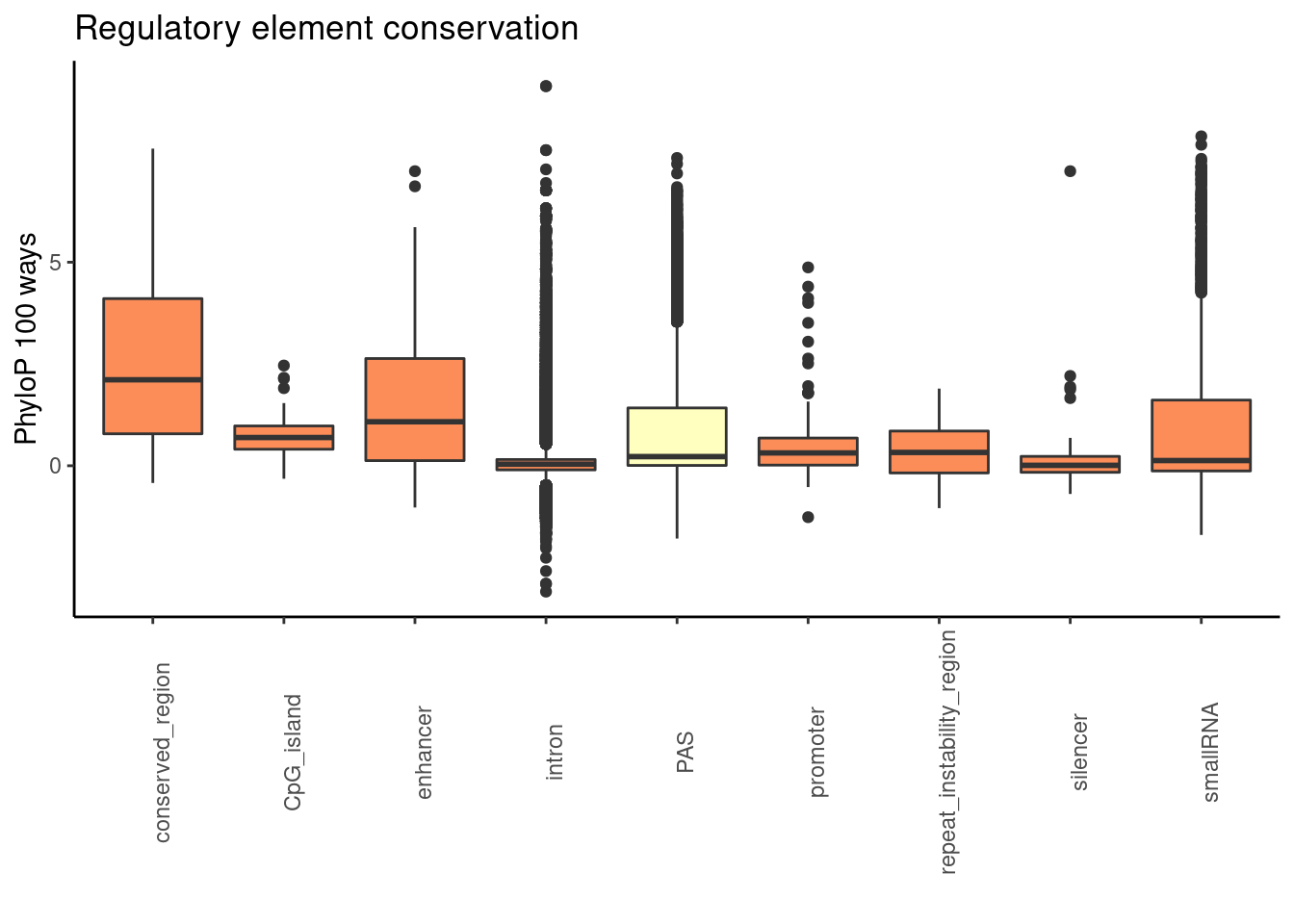
| Version | Author | Date |
|---|---|---|
| f2e7ead | brimittleman | 2020-12-24 |
promoter100= RegandRNA_100 %>% filter(element=="promoter")
enhan100= RegandRNA_100 %>% filter(element=="enhancer")
consv100= RegandRNA_100 %>% filter(element=="conserved_region")
cpg100= RegandRNA_100 %>% filter(element=="CpG_island")
repeate100= RegandRNA_100 %>% filter(element=="repeat_instability_region")
silencer100= RegandRNA_100 %>% filter(element=="silencer")
smallRNA100= RegandRNA_100 %>% filter(element=="smallRNA")
wilcox.test(PAS_100$phyloP100,promoter100$phyloP100, alternative = "greater")
Wilcoxon rank sum test with continuity correction
data: PAS_100$phyloP100 and promoter100$phyloP100
W = 5533420, p-value = 0.01534
alternative hypothesis: true location shift is greater than 0wilcox.test(PAS_100$phyloP100,enhan100$phyloP100, alternative = "greater")
Wilcoxon rank sum test with continuity correction
data: PAS_100$phyloP100 and enhan100$phyloP100
W = 22820110, p-value = 1
alternative hypothesis: true location shift is greater than 0wilcox.test(PAS_100$phyloP100,consv100$phyloP100, alternative = "greater")
Wilcoxon rank sum test with continuity correction
data: PAS_100$phyloP100 and consv100$phyloP100
W = 1046316, p-value = 1
alternative hypothesis: true location shift is greater than 0wilcox.test(PAS_100$phyloP100,cpg100$phyloP100, alternative = "greater")
Wilcoxon rank sum test with continuity correction
data: PAS_100$phyloP100 and cpg100$phyloP100
W = 2135792, p-value = 0.9998
alternative hypothesis: true location shift is greater than 0wilcox.test(PAS_100$phyloP100,repeate100$phyloP100, alternative = "greater")
Wilcoxon rank sum test with continuity correction
data: PAS_100$phyloP100 and repeate100$phyloP100
W = 1138733, p-value = 0.0201
alternative hypothesis: true location shift is greater than 0wilcox.test(PAS_100$phyloP100,silencer100$phyloP100, alternative = "greater")
Wilcoxon rank sum test with continuity correction
data: PAS_100$phyloP100 and silencer100$phyloP100
W = 1407416, p-value = 6.544e-06
alternative hypothesis: true location shift is greater than 0wilcox.test(PAS_100$phyloP100,smallRNA100$phyloP100, alternative = "greater")
Wilcoxon rank sum test with continuity correction
data: PAS_100$phyloP100 and smallRNA100$phyloP100
W = 54968724, p-value < 2.2e-16
alternative hypothesis: true location shift is greater than 0Map the promoters to genes and subset to genes we have APA in.
The regulatory elements is a really small set…
regelements=read.table("../data/RegElements/hg38_RefSeqElements.bed", stringsAsFactors = F, col.names = c("chr","start","end", "type", "score", "strand","hardsoft", "hardend","color" ))
promoters=regelements %>% filter(type=="promoter", chr !="chrX", chr!="chrY") %>% select(-hardsoft, -hardend, -color)
nrow(promoters)[1] 230write.table(promoters, "../data/RegElements/hg38_promoters.bed", col.names = F, row.names = F, quote = F,sep="\t")Map these to genes:
#bedtools map -a ../data/cleanPeaks_lifted/AllPAS_postLift.sort.bed -b ../../genome_anotation_data/hg38_refseq_anno/hg38_ncbiRefseq_Formatted_Allannotation_noSNO.Resort.bed -c 4 -S -o distinct > ../data/cleanPeaks_anno/AllPAS_postLift.sort_LocAnno.bed
sort -k1,1 -k2,2n ../data/RegElements/hg38_promoters.bed > ../data/RegElements/hg38_promoters_sort.bed
sort -k1,1 -k2,2n ../../genome_anotation_data/hg38_refseq_anno/hg38_ncbiRefseq_GenesParsed.bed > ../data/RegElements/hg38_ncbiRefseq_GenesParsed_sort.bed
bedtools map -a ../data/RegElements/hg38_promoters_sort.bed -b ../data/RegElements/hg38_ncbiRefseq_GenesParsed_sort.bed -c 4 -s -o distinct > ../data/RegElements/hg38_promoters_genes.bedPromoters with genes:
promgene=read.table("../data/RegElements/hg38_promoters_genes.bed",col.names = c("chr", "start", "end", "element", "score", "strand","gene"),stringsAsFactors = F)assign to genes and filter to genes in the PAS set
genesPAS=read.table("../data/PAS_doubleFilter/PAS_5perc_either_HumanCoord_BothUsage_meta_doubleFilter.txt",header=T,stringsAsFactors = F) %>% select(gene) %>% unique()
Reg_20Promotergene= read.table("../data/RegElements/hg38_RefSeqElements.phyloP20.txt", col.names = c('chr', 'start', 'end', 'element', 'phyloP20' ), header = F, stringsAsFactors = F) %>% inner_join(promgene,by=c("chr", "start", "end", "element")) %>% filter(gene %in% genesPAS$gene)
Reg_100Promotergene= read.table("../data/RegElements/hg38_RefSeqElements.phyloP100.txt", col.names = c('chr', 'start', 'end', 'element', 'phyloP20' ), header = F, stringsAsFactors = F) %>% inner_join(promgene,by=c("chr", "start", "end", "element"))%>% filter(gene %in% genesPAS$gene)Too few to use.
Define promoter region at 250 upstream of the PAS.
AllgeneProm=read.table("../data/RegElements/hg38_ncbiRefseq_GenesParsed_sort.bed", col.names = c("chr", "start", "end", "gene","score", "strand")) %>% filter(gene %in% genesPAS$gene) %>% mutate(PromStart=ifelse(strand=="+", start-200, end), PromEnd=ifelse(strand=="+", start, end+200)) %>% select(chr, PromStart, PromEnd, gene, score, strand)
PromAPAgenes=write.table(AllgeneProm, "../data/RegElements/PromGenes.bed",col.names = F, row.names = F, quote = F, sep="\t")python extractPhylopGeneral_20way.py ../data/RegElements/PromGenes.bed ../data/RegElements/hg38_PromGenes.P20.txt
python extractPhylopGeneral.py ../data/RegElements/PromGenes.bed ../data/RegElements/hg38_PromGenes.P100.txtProm_wPAS_20=read.table("../data/RegElements/hg38_PromGenes.P20.txt", col.names = c("chr","start","end","PAS", "phyloP20"), stringsAsFactors = F) %>% drop_na() %>% mutate(element="Promoter") %>% select(element,phyloP20) %>% bind_rows(PAS_20)
Prom_wPAS_100=read.table("../data/PhyloP/PAS_phyloP.txt", col.names = c("chr","start","end", "phyloP100"), stringsAsFactors = F) %>% drop_na() %>% mutate(element="Promoter") %>% select(element,phyloP100) %>% bind_rows(PAS_100)ggplot(Prom_wPAS_100,aes(x=element,y=phyloP100)) + geom_boxplot() + labs(title="PAS vs. promoters for 100 way Phylop")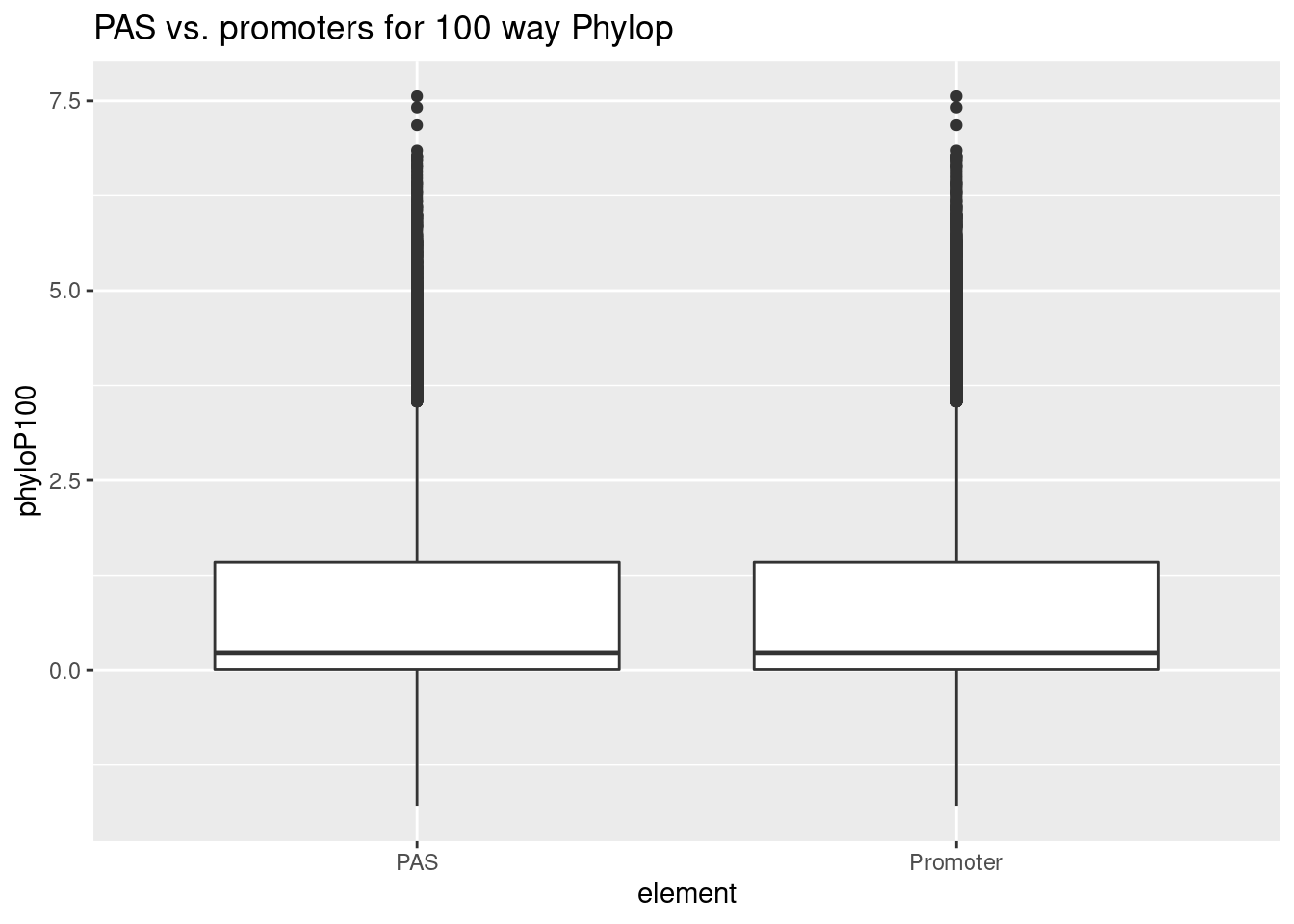
ggplot(Prom_wPAS_20,aes(x=element,y=phyloP20)) + geom_boxplot() + labs(title="PAS vs. promoters for 20 way Phylop")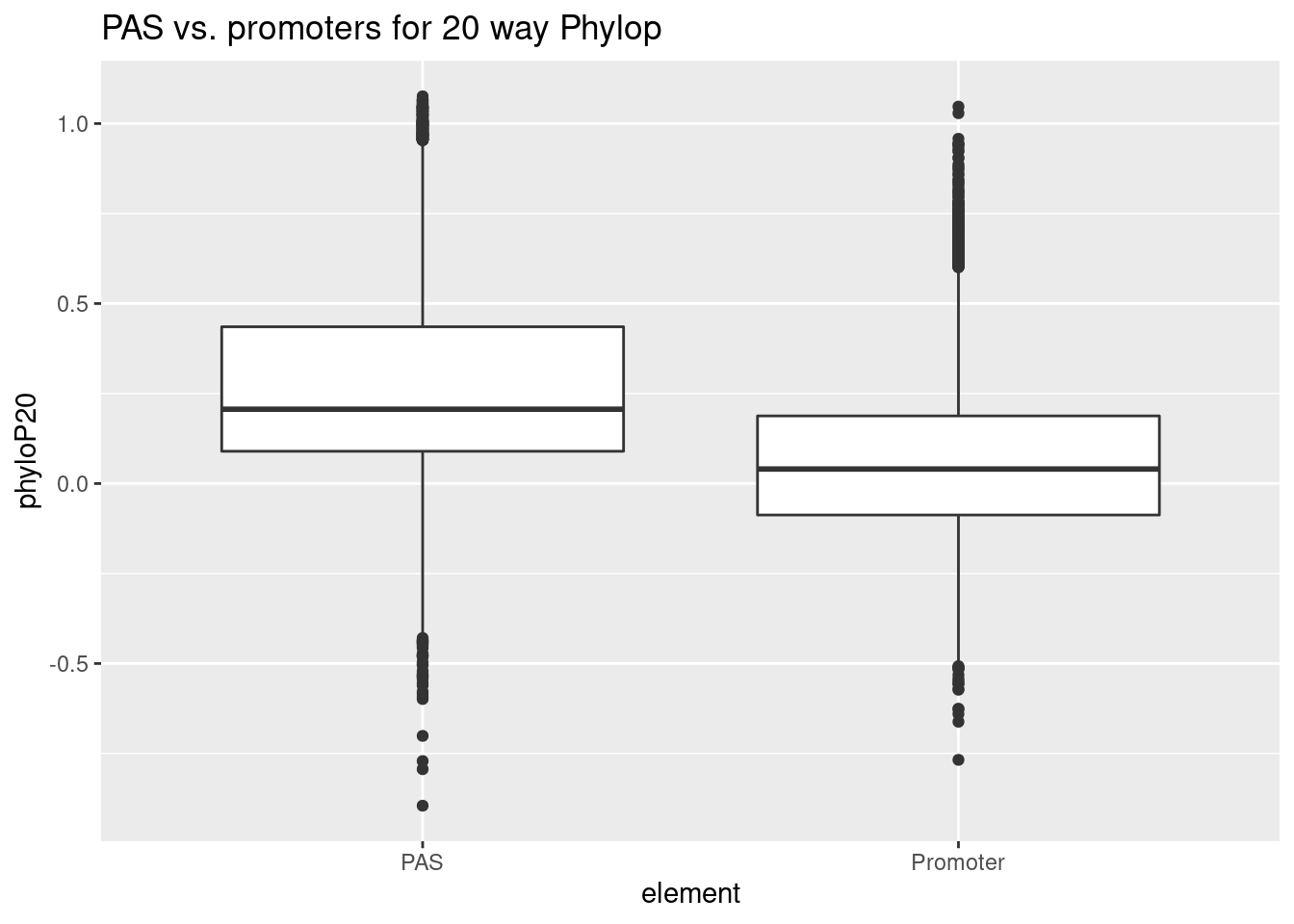
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.4.0 stringr_1.4.0 dplyr_0.8.3 purrr_0.3.4
[5] readr_1.3.1 tidyr_1.1.0 tibble_2.1.3 ggplot2_3.2.1
[9] tidyverse_1.3.0 cowplot_1.0.0 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.5 lubridate_1.7.4 lattice_0.20-38
[4] assertthat_0.2.1 rprojroot_1.3-2 digest_0.6.20
[7] R6_2.4.0 cellranger_1.1.0 backports_1.1.4
[10] reprex_0.3.0 evaluate_0.14 httr_1.4.1
[13] pillar_1.4.2 rlang_0.4.6 lazyeval_0.2.2
[16] readxl_1.3.1 rstudioapi_0.10 whisker_0.3-2
[19] Matrix_1.2-18 reticulate_1.16 rmarkdown_1.13
[22] labeling_0.3 munsell_0.5.0 broom_0.5.2
[25] compiler_3.6.1 httpuv_1.5.1 modelr_0.1.8
[28] xfun_0.8 pkgconfig_2.0.2 htmltools_0.3.6
[31] tidyselect_1.1.0 fansi_0.4.0 crayon_1.3.4
[34] dbplyr_1.4.2 withr_2.1.2 later_0.8.0
[37] grid_3.6.1 nlme_3.1-140 jsonlite_1.6
[40] gtable_0.3.0 lifecycle_0.1.0 DBI_1.1.0
[43] git2r_0.26.1 magrittr_1.5 scales_1.1.0
[46] cli_2.2.0 stringi_1.4.3 farver_2.0.1
[49] fs_1.3.1 promises_1.0.1 xml2_1.3.2
[52] ellipsis_0.2.0.1 vctrs_0.3.0 generics_0.0.2
[55] RColorBrewer_1.1-2 tools_3.6.1 glue_1.3.1
[58] hms_0.5.3 yaml_2.2.0 colorspace_1.4-1
[61] rvest_0.3.5 knitr_1.23 haven_2.3.1