PAS descriptive plots
Briana Mittleman
4/23/2019
Last updated: 2019-09-04
Checks: 6 1
Knit directory: apaQTL/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.4.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
The global environment had objects present when the code in the R Markdown file was run. These objects can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment. Use wflow_publish or wflow_build to ensure that the code is always run in an empty environment.
The following objects were defined in the global environment when these results were created:
| Name | Class | Size |
|---|---|---|
| data | environment | 56 bytes |
| env | environment | 56 bytes |
The command set.seed(20190411) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/.DS_Store
Ignored: data/mergedBW_byfrac/
Ignored: docs/.DS_Store
Ignored: docs/figure/.DS_Store
Ignored: output/.DS_Store
Untracked files:
Untracked: .Rprofile
Untracked: ._.DS_Store
Untracked: .gitignore
Untracked: @
Untracked: _workflowr.yml
Untracked: analysis/._PASdescriptiveplots.Rmd
Untracked: analysis/._cuttoffPercUsage.Rmd
Untracked: analysis/QTLexampleplots.Rmd
Untracked: analysis/cuttoffPercUsage.Rmd
Untracked: analysis/eQTLoverlap.Rmd
Untracked: analysis/interpret verify bam.Rmd
Untracked: analysis/interpret_verifybam.Rmd
Untracked: analysis/mergeRNA.Rmd
Untracked: analysis/oldstuffNotNeeded.Rmd
Untracked: analysis/remove_badlines.Rmd
Untracked: analysis/totalspec.Rmd
Untracked: apaQTL.Rproj
Untracked: code/.NascentRNAdtPlotFirstintronicPAS.sh.swp
Untracked: code/._ApaQTL_nominalNonnorm.sh
Untracked: code/._BothFracDTPlotGeneRegions.sh
Untracked: code/._BothFracDTPlotGeneRegions_normalized.sh
Untracked: code/._EandPqtl_perm.sh
Untracked: code/._EandPqtls.sh
Untracked: code/._FC_NucintornUpandDown.sh
Untracked: code/._FC_UTR.sh
Untracked: code/._FC_intornUpandDownsteamPAS.sh
Untracked: code/._FC_nascentseq.sh
Untracked: code/._FC_newPeaks_olddata.sh
Untracked: code/._HMMpermuteTotal.py
Untracked: code/._HmmPermute.py
Untracked: code/._IntronicPASDT.sh
Untracked: code/._LC_samplegroups.py
Untracked: code/._LD_qtl.sh
Untracked: code/._LD_snpsproxy.sh
Untracked: code/._NascentRNAdtPlot.sh
Untracked: code/._NascentRNAdtPlot3UTRPAS.sh
Untracked: code/._NascentRNAdtPlotExcludeFirstintronicPAS.sh
Untracked: code/._NascentRNAdtPlotNucPAS.sh
Untracked: code/._NascentRNAdtPlotTotPAS.sh
Untracked: code/._NascentRNAdtPlotintronicPAS.sh
Untracked: code/._NascnetRNAdtPlotPAS.sh
Untracked: code/._NetSeq_fourthintronDT.sh
Untracked: code/._NomResfromPASSNP.py
Untracked: code/._NuclearPAS_5per.bed.py
Untracked: code/._PTTfacetboxplots.R
Untracked: code/._PrematureQTLNominal.sh
Untracked: code/._PrematureQTLPermuted.sh
Untracked: code/._QTL2bed.py
Untracked: code/._QTL2bed_withstrand.py
Untracked: code/._RNAbam2bw.sh
Untracked: code/._RNAseqDTplot.sh
Untracked: code/._SAF215upbed.py
Untracked: code/._SnakefilePAS
Untracked: code/._SnakefilefiltPAS
Untracked: code/._TESplots100bp.sh
Untracked: code/._TESplots150bp.sh
Untracked: code/._TESplots200bp.sh
Untracked: code/._TotalPAS_5perc.bed.py
Untracked: code/._Untitled
Untracked: code/._ZipandTabPheno.sh
Untracked: code/._aAPAqtl_nominal39ind.sh
Untracked: code/._annotatePacBioPASregion.sh
Untracked: code/._annotatedPAS2bed.py
Untracked: code/._apaInPandE.py
Untracked: code/._apaQTLCorrectPvalMakeQQ.R
Untracked: code/._apaQTLCorrectpval_6or7a.R
Untracked: code/._apaQTL_Nominal.sh
Untracked: code/._apaQTL_nominalv67.sh
Untracked: code/._apaQTL_permuted.sh
Untracked: code/._apaQTL_permuted_test6A7A.sh
Untracked: code/._apainRibo.py
Untracked: code/._assignNucIntonpeak2intronlocs.sh
Untracked: code/._assignTotIntronpeak2intronlocs.sh
Untracked: code/._bam2BW_5primemost.sh
Untracked: code/._bed2saf.py
Untracked: code/._bothFracDTplot1stintron.sh
Untracked: code/._bothFracDTplot4thintron.sh
Untracked: code/._bothFrac_FC.sh
Untracked: code/._callPeaksYL.py
Untracked: code/._changeRibonomQTLres2genename.py
Untracked: code/._changenomQTLres2geneName.py
Untracked: code/._chooseAnno2PAS_pacbio.py
Untracked: code/._chooseAnno2SAF.py
Untracked: code/._chooseSignalSite
Untracked: code/._chooseSignalSite.py
Untracked: code/._closestannotated.sh
Untracked: code/._closestannotated_byfrac.sh
Untracked: code/._cluster.json
Untracked: code/._clusterPAS.json
Untracked: code/._clusterfiltPAS.json
Untracked: code/._codingdms2bed.py
Untracked: code/._config.yaml
Untracked: code/._config2.yaml
Untracked: code/._configOLD.yaml
Untracked: code/._convertNominal2SNPLOC.py
Untracked: code/._convertNominal2SNPloc2Versions.py
Untracked: code/._convertNumeric.py
Untracked: code/._correctNomeqtl.R
Untracked: code/._createPlinkSampfile.py
Untracked: code/._dag.pdf
Untracked: code/._eQTL_switch2snploc.py
Untracked: code/._eQTLgenestestedapa.py
Untracked: code/._encodeRNADTplots.sh
Untracked: code/._extractGenotypes.py
Untracked: code/._extractseqfromqtlfastq.py
Untracked: code/._fc2leafphen.py
Untracked: code/._fc_filteredPAS6and7As.sh
Untracked: code/._fifteenBPupstreamPAS.py
Untracked: code/._fiftyBPupstreamPAS.py
Untracked: code/._filter5perc.R
Untracked: code/._filter5percPheno.py
Untracked: code/._filterLDsnps.py
Untracked: code/._filterMPPAS.py
Untracked: code/._filterMPPAS_15.py
Untracked: code/._filterMPPAS_15_7As.py
Untracked: code/._filterMPPAS_50.py
Untracked: code/._filterSAFforMP.py
Untracked: code/._filterpeaks.py
Untracked: code/._finalPASbed2SAF.py
Untracked: code/._fix4su304corr.py
Untracked: code/._fix4su604corr.py
Untracked: code/._fix4sukalisto.py
Untracked: code/._fixExandUnexeQTL
Untracked: code/._fixExandUnexeQTL.py
Untracked: code/._fixFChead.py
Untracked: code/._fixFChead_bothfrac.py
Untracked: code/._fixFChead_short.py
Untracked: code/._fixH3k12ac.py
Untracked: code/._fixPASregionSNPs.py
Untracked: code/._fixRNAhead4corr.py
Untracked: code/._fixRNAkalisto.py
Untracked: code/._fixgroupedtranscript.py
Untracked: code/._fixhead_netseqfc.py
Untracked: code/._getAPAfromanyeQTL.py
Untracked: code/._getApapval4eqtl.py
Untracked: code/._getApapval4eqtl_unexp.py
Untracked: code/._getApapval4eqtl_version67.py
Untracked: code/._getDownstreamIntronNuclear.py
Untracked: code/._getIntronDownstreamPAS.py
Untracked: code/._getIntronUpstreamPAS.py
Untracked: code/._getQTLalleles.py
Untracked: code/._getQTLfastq.sh
Untracked: code/._getUpstreamIntronNuclear.py
Untracked: code/._grouptranscripts.py
Untracked: code/._intersectVCFandupPAS.sh
Untracked: code/._keep5perMAF.py
Untracked: code/._keepSNP_vcf.sh
Untracked: code/._make5percPeakbed.py
Untracked: code/._makeFileID.py
Untracked: code/._makePheno.py
Untracked: code/._makeSAFbothfrac5perc.py
Untracked: code/._makeSNP2rsidfile.py
Untracked: code/._makeeQTLempirical_unexp.py
Untracked: code/._makeeQTLempiricaldist.py
Untracked: code/._makegencondeTSSfile.py
Untracked: code/._mapSSsnps2PAS.sh
Untracked: code/._mergRNABam.sh
Untracked: code/._mergeAllBam.sh
Untracked: code/._mergeBW_norm.sh
Untracked: code/._mergeBamNascent.sh
Untracked: code/._mergeByFracBam.sh
Untracked: code/._mergePeaks.sh
Untracked: code/._mnase1stintron.sh
Untracked: code/._mnaseDT_fourthintron.sh
Untracked: code/._namePeaks.py
Untracked: code/._netseqDTplot1stIntron.sh
Untracked: code/._netseqFC.sh
Untracked: code/._nucQTLGWAS.py
Untracked: code/._nucSpecQTLineData.py
Untracked: code/._nucSpeceffectsize.py
Untracked: code/._pQTLsotherdata.py
Untracked: code/._pacbioDT.sh
Untracked: code/._pacbioIntronicDT.sh
Untracked: code/._parseBestbamid.py
Untracked: code/._peak2PAS.py
Untracked: code/._peakFC.sh
Untracked: code/._pheno2countonly.R
Untracked: code/._phenoQTLfromlist.py
Untracked: code/._processYRIgen.py
Untracked: code/._pttQTLsinapaQTL.py
Untracked: code/._qtlRegionseq.sh
Untracked: code/._qtlsPvalOppFrac.py
Untracked: code/._quantassign2parsedpeak.py
Untracked: code/._removeXfromHmm.py
Untracked: code/._removeloc_pheno.py
Untracked: code/._riboQTL.sh
Untracked: code/._runCorrectNomEqtl.sh
Untracked: code/._runHMMpermuteAPAqtls.sh
Untracked: code/._runHMMpermuteeQTLS.sh
Untracked: code/._runMakeEmpiricaleQTL_unexp.sh
Untracked: code/._runMakeeQTLempirical.sh
Untracked: code/._run_bam2bw_all3prime.sh
Untracked: code/._run_bam2bw_extra3.sh
Untracked: code/._run_bestbamid.sj
Untracked: code/._run_filtersnpLD.sh
Untracked: code/._run_getAPAfromeQTL_version6.7.sh
Untracked: code/._run_getApaPval4eqtl.sh
Untracked: code/._run_getapafromeQTL.py
Untracked: code/._run_getapafromeQTL.sh
Untracked: code/._run_getapapval4eqtl_unexp.sh
Untracked: code/._run_leafcutterDiffIso.sh
Untracked: code/._run_prxySNP.sh
Untracked: code/._run_pttfacetboxplot.sh
Untracked: code/._run_sepUsagephen.sh
Untracked: code/._run_sepgenobychrom.sh
Untracked: code/._run_verifybam.sh
Untracked: code/._selectNominalPvalues.py
Untracked: code/._sepUsagePhen.py
Untracked: code/._sepgenobychrom.py
Untracked: code/._snakemakePAS.batch
Untracked: code/._snakemakefiltPAS.batch
Untracked: code/._sortindexRNAbam.sh
Untracked: code/._submit-snakemakePAS.sh
Untracked: code/._submit-snakemakefiltPAS.sh
Untracked: code/._subsetAPAnotEorPgene.py
Untracked: code/._subsetAPAnotEorPgene_2versions.py
Untracked: code/._subsetApanoteGene.py
Untracked: code/._subsetApanoteGene_2versions.py
Untracked: code/._subsetUnexplainedeQTLs.py
Untracked: code/._subsetVCF_SS.sh
Untracked: code/._subsetVCF_noSSregions.sh
Untracked: code/._subsetVCF_upstreamPAS.sh
Untracked: code/._subset_diffisopheno.py
Untracked: code/._subsetpermAPAwithGenelist.py
Untracked: code/._subsetpermAPAwithGenelist_2versions.py
Untracked: code/._subsetvcf_otherreg.sh
Untracked: code/._subsetvcf_permSS.sh
Untracked: code/._subtrachfiveprimeUTR.sh
Untracked: code/._subtractExons.sh
Untracked: code/._subtractfiveprimeUTR.sh
Untracked: code/._tabixSNPS.sh
Untracked: code/._tenBPupstreamPAS.py
Untracked: code/._testVerifyBam.sh
Untracked: code/._totSeceffectsize.py
Untracked: code/._twentyBPupstreamPAS.py
Untracked: code/._utrdms2saf.py
Untracked: code/._vcf2bed.py
Untracked: code/._verifyBam18517N.sh
Untracked: code/._verifyBam18517T.sh
Untracked: code/._verifyBam19128N.sh
Untracked: code/._verifyBam19128T.sh
Untracked: code/._wrap_verifybam.sh
Untracked: code/._writePTTexamplecode.py
Untracked: code/._writePTTexamplecode.sh
Untracked: code/.pversion
Untracked: code/.snakemake/
Untracked: code/1
Untracked: code/APAqtl_nominal.err
Untracked: code/APAqtl_nominal.out
Untracked: code/APAqtl_nominal_39.err
Untracked: code/APAqtl_nominal_39.out
Untracked: code/APAqtl_nominal_nonNorm.err
Untracked: code/APAqtl_nominal_nonNorm.out
Untracked: code/APAqtl_nominal_versions67.err
Untracked: code/APAqtl_nominal_versions67.out
Untracked: code/APAqtl_permuted.err
Untracked: code/APAqtl_permuted.out
Untracked: code/APAqtl_permuted_versions67.err
Untracked: code/APAqtl_permuted_versions67.out
Untracked: code/ApaQTL_nominalNonnorm.sh
Untracked: code/BothFracDTPlot1stintron.err
Untracked: code/BothFracDTPlot1stintron.out
Untracked: code/BothFracDTPlot4stintron.err
Untracked: code/BothFracDTPlot4stintron.out
Untracked: code/BothFracDTPlotGeneRegions.err
Untracked: code/BothFracDTPlotGeneRegions.out
Untracked: code/BothFracDTPlotGeneRegions_norm.err
Untracked: code/BothFracDTPlotGeneRegions_norm.out
Untracked: code/BothFracDTPlotGeneRegions_normalized.sh
Untracked: code/DistPAS2Sig.py
Untracked: code/EandPqtl.err
Untracked: code/EandPqtl.out
Untracked: code/EandPqtl_perm.sh
Untracked: code/EandPqtls.sh
Untracked: code/EncodeRNADTPlotGeneRegions.err
Untracked: code/EncodeRNADTPlotGeneRegions.out
Untracked: code/FC_NucintornUpandDown.sh
Untracked: code/FC_NucintronPASupandDown.err
Untracked: code/FC_NucintronPASupandDown.out
Untracked: code/FC_UTR.err
Untracked: code/FC_UTR.out
Untracked: code/FC_UTR.sh
Untracked: code/FC_intornUpandDownsteamPAS.sh
Untracked: code/FC_intronPASupandDown.err
Untracked: code/FC_intronPASupandDown.out
Untracked: code/FC_nascent.err
Untracked: code/FC_nascentout
Untracked: code/FC_nascentseq.sh
Untracked: code/FC_newPAS_olddata.err
Untracked: code/FC_newPAS_olddata.out
Untracked: code/FC_newPeaks_olddata.sh
Untracked: code/HMMpermuteTotal.py
Untracked: code/HmmPermute.p
Untracked: code/HmmPermute.py
Untracked: code/IntronicPASDT.err
Untracked: code/IntronicPASDT.out
Untracked: code/IntronicPASDT.sh
Untracked: code/LC_samplegroups.py
Untracked: code/LD_qtl.sh
Untracked: code/LD_snpsproxy.sh
Untracked: code/LD_vcftools.hap.out
Untracked: code/NascentDTPlotGeneRegions.err
Untracked: code/NascentDTPlotGeneRegions.out
Untracked: code/NascentDTPlotPAS.err
Untracked: code/NascentDTPlotPAS.out
Untracked: code/NascentDTPlotPAS_3utr.err
Untracked: code/NascentDTPlotPAS_3utr.out
Untracked: code/NascentDTPlotPAS_firstintron.err
Untracked: code/NascentDTPlotPAS_firstintron.out
Untracked: code/NascentDTPlotPAS_intron.err
Untracked: code/NascentDTPlotPAS_intron.out
Untracked: code/NascentDTPlotPAS_nuc.err
Untracked: code/NascentDTPlotPAS_nuc.out
Untracked: code/NascentDTPlotPAS_tot.err
Untracked: code/NascentDTPlotPAS_tot.out
Untracked: code/NascentRNAdtPlot.sh
Untracked: code/NascentRNAdtPlot3UTRPAS.sh
Untracked: code/NascentRNAdtPlotExcludeFirstintronicPAS.sh
Untracked: code/NascentRNAdtPlotFirstintronicPAS.sh
Untracked: code/NascentRNAdtPlotNucPAS.sh
Untracked: code/NascentRNAdtPlotTotPAS.sh
Untracked: code/NascentRNAdtPlotintronicPAS.sh
Untracked: code/NascnetRNAdtPlotPAS.sh
Untracked: code/NetSeq_fourthintronDT.sh
Untracked: code/NomResfromPASSNP.py
Untracked: code/NuclearPAS_5per.bed.py
Untracked: code/Nuclear_example.err
Untracked: code/Nuclear_example.out
Untracked: code/PACbioDT.err
Untracked: code/PACbioDT.out
Untracked: code/PACbioDTitronic.err
Untracked: code/PACbioDTitronic.out
Untracked: code/PTTfacetboxplots.R
Untracked: code/PrematureQTLNominal.sh
Untracked: code/PrematureQTLPermuted.sh
Untracked: code/Prematureqtl_nominal.err
Untracked: code/Prematureqtl_nominal.out
Untracked: code/Prematureqtl_permuted.err
Untracked: code/Prematureqtl_permuted.out
Untracked: code/QTL2bed.py
Untracked: code/QTL2bed_withstrand.py
Untracked: code/README.md
Untracked: code/RNABam2BW.err
Untracked: code/RNABam2BW.out
Untracked: code/RNAbam2bw.sh
Untracked: code/RNAseqDTPlotGeneRegions.err
Untracked: code/RNAseqDTPlotGeneRegions.out
Untracked: code/RNAseqDTplot.sh
Untracked: code/Rplots.pdf
Untracked: code/SAF215upbed.py
Untracked: code/SAF215upbed_gen.py
Untracked: code/Script4NuclearPTTqtlexamples.sh
Untracked: code/Script4NuclearQTLexamples.sh
Untracked: code/Script4TotalPTTqtlexamples.sh
Untracked: code/Script4TotalQTLexamples.sh
Untracked: code/TESplots100bp.err
Untracked: code/TESplots100bp.out
Untracked: code/TESplots100bp.sh
Untracked: code/TESplots150bp.err
Untracked: code/TESplots150bp.out
Untracked: code/TESplots150bp.sh
Untracked: code/TESplots200bp.err
Untracked: code/TESplots200bp.out
Untracked: code/TESplots200bp.sh
Untracked: code/TotalPAS_5perc.bed.py
Untracked: code/Total_example.err
Untracked: code/Total_example.out
Untracked: code/Untitled
Untracked: code/Upstream100Bases_general.py
Untracked: code/YRI_LCL.vcf.gz
Untracked: code/YRI_LCL_chr1.vcf.gz.log
Untracked: code/YRI_LCL_chr1.vcf.gz.recode.vcf
Untracked: code/ZipandTabPheno.sh
Untracked: code/aAPAqtl_nominal39ind.sh
Untracked: code/annotatePacBioPASregion.sh
Untracked: code/annotatedPAS2bed.py
Untracked: code/annotatedPASregion.err
Untracked: code/annotatedPASregion.out
Untracked: code/apaInPandE.py
Untracked: code/apaQTLCorrectPvalMakeQQ_4pc.R
Untracked: code/apaQTLCorrectpval_6or7a.R
Untracked: code/apaQTL_Nominal_4pc.sh
Untracked: code/apaQTL_nominalv67.sh
Untracked: code/apaQTL_permuted.4pc.sh
Untracked: code/apaQTL_permuted_test6A7A.sh
Untracked: code/apafacetboxplots.R
Untracked: code/apainRibo.py
Untracked: code/apaqtlfacetboxplots.R
Untracked: code/assignNucIntonpeak2intronlocs.sh
Untracked: code/assignPeak2Intronicregion.err
Untracked: code/assignPeak2Intronicregion.out
Untracked: code/assignTotIntronpeak2intronlocs.sh
Untracked: code/assigntotPeak2Intronicregion.err
Untracked: code/assigntotPeak2Intronicregion.out
Untracked: code/bam2BW_5primemost.sh
Untracked: code/bam2bw.err
Untracked: code/bam2bw.out
Untracked: code/bam2bw_5primemost.err
Untracked: code/bam2bw_5primemost.out
Untracked: code/binary_fileset.log
Untracked: code/bothFracDTplot1stintron.sh
Untracked: code/bothFracDTplot4thintron.sh
Untracked: code/bothFrac_FC.err
Untracked: code/bothFrac_FC.out
Untracked: code/bothFrac_FC.sh
Untracked: code/callSHscripts.txt
Untracked: code/changePermQTLres2geneName.py
Untracked: code/changeRibonomQTLres2genename.py
Untracked: code/changenomQTLres2geneName.py
Untracked: code/chooseAnno2PAS_pacbio.py
Untracked: code/closestannotated.err
Untracked: code/closestannotated.out
Untracked: code/closestannotated.sh
Untracked: code/closestannotated_byfrac.sh
Untracked: code/closestannotatedbyfrac.err
Untracked: code/closestannotatedbyfrac.out
Untracked: code/codingdms2bed.py
Untracked: code/convertNominal2SNPLOC.py
Untracked: code/convertNominal2SNPloc2Versions.py
Untracked: code/correctNomeqtl.R
Untracked: code/createPlinkSampfile.py
Untracked: code/dag.pdf
Untracked: code/dagPAS.pdf
Untracked: code/dagfiltPAS.pdf
Untracked: code/eQTL_switch2snploc.py
Untracked: code/eQTLgenestestedapa.py
Untracked: code/encodeRNADTplots.sh
Untracked: code/environmentLeaf.yaml
Untracked: code/extractGenotypes.py
Untracked: code/extractseqfromqtlfastq.py
Untracked: code/fc2leafphen.py
Untracked: code/fc_filteredPAS6and7As.sh
Untracked: code/fifteenBPupstreamPAS.py
Untracked: code/fiftyBPupstreamPAS.py
Untracked: code/filterLDsnps.py
Untracked: code/filterMPPAS.py
Untracked: code/filterMPPAS_15.py
Untracked: code/filterMPPAS_15_7As.py
Untracked: code/filterMPPAS_50.py
Untracked: code/filterSAFforMP.py
Untracked: code/filterSAFforMP_gen.py
Untracked: code/finalPASbed2SAF.py
Untracked: code/findbuginpeaks.R
Untracked: code/fix4su304corr.py
Untracked: code/fix4su604corr.py
Untracked: code/fix4sukalisto.py
Untracked: code/fixExandUnexeQTL
Untracked: code/fixExandUnexeQTL.py
Untracked: code/fixFChead_bothfrac.py
Untracked: code/fixFChead_short.py
Untracked: code/fixFChead_summary.py
Untracked: code/fixH3k12ac.py
Untracked: code/fixPASregionSNPs.py
Untracked: code/fixRNAhead4corr.py
Untracked: code/fixRNAkalisto.py
Untracked: code/fixgroupedtranscript.py
Untracked: code/fixhead_netseqfc.py
Untracked: code/genotypesYRI.gen.proc.keep.vcf.log
Untracked: code/genotypesYRI.gen.proc.keep.vcf.recode.vcf
Untracked: code/get100upPAS.py
Untracked: code/getAPAfromanyeQTL.py
Untracked: code/getApapval4eqtl.py
Untracked: code/getApapval4eqtl_unexp.py
Untracked: code/getApapval4eqtl_version67.py
Untracked: code/getDownstreamIntronNuclear.py
Untracked: code/getIntronDownstreamPAS.py
Untracked: code/getIntronUpstreamPAS.py
Untracked: code/getQTLalleles.py
Untracked: code/getQTLfastq.sh
Untracked: code/getSeq100up.sh
Untracked: code/getUpstreamIntronNuclear.py
Untracked: code/getseq100up.err
Untracked: code/getseq100up.out
Untracked: code/grouptranscripts.err
Untracked: code/grouptranscripts.out
Untracked: code/grouptranscripts.py
Untracked: code/intersectPAS_ssSNPS.err
Untracked: code/intersectPAS_ssSNPS.out
Untracked: code/intersectVCFPAS.err
Untracked: code/intersectVCFPAS.out
Untracked: code/intersectVCFandupPAS.sh
Untracked: code/keep5perMAF.py
Untracked: code/keepSNP_vcf.sh
Untracked: code/log/
Untracked: code/makeSAFbothfrac5perc.py
Untracked: code/makeSNP2rsidfile.py
Untracked: code/makeeQTLempirical_unexp.py
Untracked: code/makeeQTLempiricaldist.py
Untracked: code/makegencondeTSSfile.py
Untracked: code/mapSSsnps2PAS.sh
Untracked: code/mergRNABam.sh
Untracked: code/mergeBW_norm.sh
Untracked: code/mergeBWnorm.err
Untracked: code/mergeBWnorm.out
Untracked: code/mergeBamNacent.err
Untracked: code/mergeBamNacent.out
Untracked: code/mergeBamNascent.sh
Untracked: code/mergeRNAbam.err
Untracked: code/mergeRNAbam.out
Untracked: code/mnase1stintron.sh
Untracked: code/mnaseDTPlot1stintron.err
Untracked: code/mnaseDTPlot1stintron.out
Untracked: code/mnaseDTPlot4thintron.err
Untracked: code/mnaseDTPlot4thintron.out
Untracked: code/mnaseDT_fourthintron.sh
Untracked: code/netDTPlot4thintron.out
Untracked: code/netseqDTplot1stIntron.sh
Untracked: code/netseqFC.err
Untracked: code/netseqFC.out
Untracked: code/netseqFC.sh
Untracked: code/neyDTPlot4thintron.err
Untracked: code/nucQTLGWAS.py
Untracked: code/nucQTLGWAS_withLD.py
Untracked: code/nucSpecQTLineData.py
Untracked: code/nucSpeceffectsize.py
Untracked: code/pQTLsotherdata.py
Untracked: code/pacbioDT.sh
Untracked: code/pacbioIntronicDT.sh
Untracked: code/parseBestbamid.py
Untracked: code/phenoQTLfromlist.py
Untracked: code/plink.log
Untracked: code/processYRIgen.py
Untracked: code/prxySNP.err
Untracked: code/prxySNP.out
Untracked: code/pttFacetBoxplots.err
Untracked: code/pttFacetBoxplots.out
Untracked: code/pttQTLsinapaQTL.py
Untracked: code/pullTwoMechData.py
Untracked: code/qtlFacetBoxplots.err
Untracked: code/qtlFacetBoxplots.out
Untracked: code/qtlRegionseq.sh
Untracked: code/qtlsPvalOppFrac.py
Untracked: code/rLD_vcftools.hap.err
Untracked: code/removeXfromHmm.py
Untracked: code/removeloc_pheno.py
Untracked: code/riboQTL.sh
Untracked: code/riboqtl.err
Untracked: code/riboqtl.out
Untracked: code/runBestBamID.err
Untracked: code/runCorrectNomEqtl.sh
Untracked: code/runCorrectNomeqtl.err
Untracked: code/runCorrectNomeqtl.out
Untracked: code/runFilterLD.err
Untracked: code/runFilterLD.out
Untracked: code/runHMMpermute.err
Untracked: code/runHMMpermute.out
Untracked: code/runHMMpermuteAPAqtls.sh
Untracked: code/runHMMpermuteeQTLS.sh
Untracked: code/runHMMpermuteeQTLs.err
Untracked: code/runHMMpermuteeQTLs.out
Untracked: code/runMakeEmpiricaleQTL_unexp.sh
Untracked: code/runMakeEmpiricaleQTLs.err
Untracked: code/runMakeEmpiricaleQTLs.out
Untracked: code/runMakeEmpiricaleQTLsunex.err
Untracked: code/runMakeEmpiricaleQTLsunex.out
Untracked: code/runMakeeQTLempirical.sh
Untracked: code/run_DistPAS2Sig.err
Untracked: code/run_DistPAS2Sig.out
Untracked: code/run_bam2bw.err
Untracked: code/run_bam2bw.out
Untracked: code/run_bam2bw_all3prime.sh
Untracked: code/run_bam2bw_extra3.sh
Untracked: code/run_bam2bwexta.err
Untracked: code/run_bam2bwexta.out
Untracked: code/run_bestbamid.sh
Untracked: code/run_distPAS2Sig.sh
Untracked: code/run_filtersnpLD.sh
Untracked: code/run_getAPAfromanyeQTL.err
Untracked: code/run_getAPAfromanyeQTL.out
Untracked: code/run_getAPAfromeQTL_version6.7.sh
Untracked: code/run_getApaPval4eQTLs.err
Untracked: code/run_getApaPval4eQTLs.out
Untracked: code/run_getApaPval4eQTLsunexplained.err
Untracked: code/run_getApaPval4eQTLsunexplained.out
Untracked: code/run_getApaPval4eqtl.sh
Untracked: code/run_getapafromeQTL.sh
Untracked: code/run_getapapval4eqtl_unexp.sh
Untracked: code/run_leafcutterDiffIso.sh
Untracked: code/run_leafcutter_ds.err
Untracked: code/run_leafcutter_ds.out
Untracked: code/run_prxySNP.sh
Untracked: code/run_pttfacetboxplot.sh
Untracked: code/run_qtlFacetBoxplots.sh
Untracked: code/run_sepUsagephen.sh
Untracked: code/run_sepgenobychrom.err
Untracked: code/run_sepgenobychrom.out
Untracked: code/run_sepgenobychrom.sh
Untracked: code/run_sepusage.err
Untracked: code/run_sepusage.out
Untracked: code/run_verifybam.err
Untracked: code/run_verifybam.out
Untracked: code/run_verifybam.sh
Untracked: code/run_verifybam128N.err
Untracked: code/run_verifybam128N.out
Untracked: code/run_verifybam128T.err
Untracked: code/run_verifybam128T.out
Untracked: code/run_verifybam517N.err
Untracked: code/run_verifybam517N.out
Untracked: code/run_verifybam517T.err
Untracked: code/run_verifybam517T.out
Untracked: code/run_verifybam_fullVCF.sh
Untracked: code/runprxySNP.err
Untracked: code/runprxySNP.out
Untracked: code/selectNominalPvalues.py
Untracked: code/sepUsagePhen.py
Untracked: code/sepgenobychrom.py
Untracked: code/seqQTLfastq.err
Untracked: code/seqQTLfastq.out
Untracked: code/seqQTLregion.err
Untracked: code/seqQTLregion.out
Untracked: code/snakePASlog.out
Untracked: code/snakefiltPASlog.out
Untracked: code/sortindexRNABam.err
Untracked: code/sortindexRNABam.out
Untracked: code/sortindexRNAbam.sh
Untracked: code/subsetAPAnotEorPgene.py
Untracked: code/subsetAPAnotEorPgene_2versions.py
Untracked: code/subsetApanoteGene.py
Untracked: code/subsetApanoteGene_2versions.py
Untracked: code/subsetUnexplainedeQTLs.py
Untracked: code/subsetVCF_SS.sh
Untracked: code/subsetVCF_noSSregions.sh
Untracked: code/subsetVCF_upstreamPAS.sh
Untracked: code/subset_diffisopheno.py
Untracked: code/subsetpermAPAwithGenelist.py
Untracked: code/subsetpermAPAwithGenelist_2versions.py
Untracked: code/subsetvcf_SS.err
Untracked: code/subsetvcf_SS.out
Untracked: code/subsetvcf_noSS.err
Untracked: code/subsetvcf_noSS.out
Untracked: code/subsetvcf_otherreg.sh
Untracked: code/subsetvcf_pas.err
Untracked: code/subsetvcf_pas.out
Untracked: code/subsetvcf_perm.err
Untracked: code/subsetvcf_perm.out
Untracked: code/subsetvcf_permSS.sh
Untracked: code/subsetvcf_rand.err
Untracked: code/subsetvcf_rand.out
Untracked: code/subtract5UTR.err
Untracked: code/subtract5UTR.out
Untracked: code/subtractExons.err
Untracked: code/subtractExons.out
Untracked: code/subtractExons.sh
Untracked: code/subtractfiveprimeUTR.sh
Untracked: code/tabixSNPS.sh
Untracked: code/tabixSNPs.err
Untracked: code/tabixSNPs.out
Untracked: code/tenBPupstreamPAS.py
Untracked: code/testVerifyBam.sh
Untracked: code/test_verifybam.err
Untracked: code/test_verifybam.out
Untracked: code/totSeceffectsize.py
Untracked: code/transcriptdm2bed.py
Untracked: code/twentyBPupstreamPAS.py
Untracked: code/utrdms2saf.py
Untracked: code/vcf2bed.py
Untracked: code/vcf_keepsnps.err
Untracked: code/vcf_keepsnps.out
Untracked: code/verifyBam18517N.sh
Untracked: code/verifyBam18517T.sh
Untracked: code/verifyBam19128N.sh
Untracked: code/verifyBam19128T.sh
Untracked: code/wrap_verifybam.err
Untracked: code/wrap_verifybam.out
Untracked: code/wrap_verifybam.sh
Untracked: code/wrap_verifybam_full.sh
Untracked: code/writeExampleQTLcode.py
Untracked: code/writePTTexamplecode.py
Untracked: code/zipandtabPhen.err
Untracked: code/zipandtabPhen.out
Untracked: data/._.DS_Store
Untracked: data/._MetaDataSequencing.txt
Untracked: data/AnnotatedPAS/
Untracked: data/ApaByEgene/
Untracked: data/ApaByPgene/
Untracked: data/BadLines/
Untracked: data/Battle_pQTL/
Untracked: data/CheckSums/
Untracked: data/CompareOldandNew/
Untracked: data/DTmatrix/
Untracked: data/DiffIso/
Untracked: data/EncodeRNA/
Untracked: data/ExampleQTLPlots/
Untracked: data/FlaggedPAS/
Untracked: data/GWAS_overlap/
Untracked: data/GeuvadisRNA/
Untracked: data/HMMqtls/
Untracked: data/Li_eQTLs/
Untracked: data/NascentRNA/
Untracked: data/NucSpeceQTLeffect/
Untracked: data/PAS/
Untracked: data/PAS_postFlag/
Untracked: data/PolyA_DB/
Untracked: data/PreTerm_pheno/
Untracked: data/PrematureQTLNominal/
Untracked: data/PrematureQTLPermuted/
Untracked: data/QTLGenotypes/
Untracked: data/QTLoverlap/
Untracked: data/QTLoverlap_nonNorm/
Untracked: data/README.md
Untracked: data/RNAseq/
Untracked: data/Reads2UTR/
Untracked: data/SNPinSS/
Untracked: data/SignalSiteFiles/
Untracked: data/TF_motifdisruption/
Untracked: data/ThirtyNineIndQtl_nominal/
Untracked: data/Version15bp6As/
Untracked: data/Version15bp7As/
Untracked: data/apaQTLNominal/
Untracked: data/apaQTLNominal_4pc/
Untracked: data/apaQTLPermuted/
Untracked: data/apaQTLPermuted_4pc/
Untracked: data/apaQTLs/
Untracked: data/assignedPeaks/
Untracked: data/assignedPeaks_15Up/
Untracked: data/bam/
Untracked: data/bam_clean/
Untracked: data/bam_waspfilt/
Untracked: data/bed_10up/
Untracked: data/bed_clean/
Untracked: data/bed_clean_sort/
Untracked: data/bed_waspfilter/
Untracked: data/bedsort_waspfilter/
Untracked: data/bothFrac_FC/
Untracked: data/bw/
Untracked: data/bw_norm/
Untracked: data/eQTLs/
Untracked: data/exampleQTLs/
Untracked: data/fastq/
Untracked: data/filterPeaks/
Untracked: data/fourSU/
Untracked: data/h3k27ac/
Untracked: data/highdiffsiggenes.txt
Untracked: data/inclusivePeaks/
Untracked: data/inclusivePeaks_FC/
Untracked: data/intronRNAratio/
Untracked: data/intron_analysis/
Untracked: data/locusZoom/
Untracked: data/mergedBG/
Untracked: data/mergedBW_norm/
Untracked: data/mergedBam/
Untracked: data/mergedbyFracBam/
Untracked: data/molPhenos/
Untracked: data/molQTLs/
Untracked: data/motifdistrupt/
Untracked: data/netseq/
Untracked: data/nonNorm_pheno/
Untracked: data/nuc_10up/
Untracked: data/nuc_10upclean/
Untracked: data/oldPASfiles/
Untracked: data/overlapeQTL_try2/
Untracked: data/overlapeQTLs/
Untracked: data/pQTLoverlap/
Untracked: data/pacbio/
Untracked: data/peakCoverage/
Untracked: data/peaks_5perc/
Untracked: data/phenotype/
Untracked: data/phenotype_5perc/
Untracked: data/pttQTL/
Untracked: data/pttQTLplots/
Untracked: data/sigDiffGenes.txt
Untracked: data/sort/
Untracked: data/sort_clean/
Untracked: data/sort_waspfilter/
Untracked: data/twoMech/
Untracked: data/verifyBAM/
Untracked: data/verifyBAM_full/
Untracked: docs/._.DS_Store
Untracked: docs/figure/._.DS_Store
Untracked: docs/figure/nonNormQTL.Rmd/._figure2D-1.pdf
Untracked: nohup.out
Untracked: output/._.DS_Store
Untracked: output/._meanCorrelationPhenotypes.svg
Untracked: output/dtPlots/
Untracked: output/fastqc/
Untracked: output/meanCorrelationPhenotypes.svg
Untracked: run_verifybam517N.err
Untracked: run_verifybam517N.out
Unstaged changes:
Modified: analysis/NuclearSpecAPAqtl.Rmd
Modified: analysis/PrematureTermQTL.Rmd
Modified: analysis/Readdistagainstfeatures.Rmd
Modified: analysis/compareAnnotatedpas.Rmd
Modified: analysis/overlapapaqtlsandeqtls.Rmd
Modified: analysis/rerunQTL_changePC.Rmd
Modified: analysis/version15bpfilter.Rmd
Modified: code/BothFracDTPlotGeneRegions.sh
Modified: code/Snakefile
Modified: code/SnakefilefiltPAS
Modified: code/apaQTLCorrectPvalMakeQQ.R
Modified: code/apaQTL_Nominal.sh
Modified: code/apaQTL_permuted.sh
Modified: code/apaQTLsnake.err
Modified: code/bam2bw.sh
Modified: code/bed2saf.py
Modified: code/cluster.json
Modified: code/clusterfiltPAS.json
Modified: code/config.yaml
Modified: code/environment.yaml
Modified: code/makePheno.py
Modified: code/mergeAllBam.sh
Modified: code/mergeByFracBam.sh
Modified: code/mergePeaks.sh
Modified: code/peakFC.sh
Modified: code/snakemake.batch
Modified: code/snakemakePAS.batch
Modified: code/snakemakefiltPAS.batch
Modified: code/submit-snakemake.sh
Modified: code/submit-snakemakePAS.sh
Modified: code/submit-snakemakefiltPAS.sh
Deleted: code/test.txt
Modified: data/MetaDataSequencing.txt
Deleted: docs/figure/PASdescriptiveplots.Rmd/figure1D-1.pdf
Deleted: reads_graphs.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 5984fad | brimittleman | 2019-09-04 | update for new filter condition |
| html | 1419181 | brimittleman | 2019-08-01 | Build site. |
| Rmd | a66cd73 | brimittleman | 2019-08-01 | fig1D pdf |
| html | dc5e7d0 | brimittleman | 2019-07-16 | Build site. |
| Rmd | 66c1c27 | brimittleman | 2019-07-16 | remove white space between bars |
| html | 96d85de | brimittleman | 2019-07-07 | Build site. |
| html | b6aa8cb | brimittleman | 2019-07-02 | Build site. |
| Rmd | d30bbd0 | brimittleman | 2019-07-02 | use only protien coding |
| html | b3cbd22 | brimittleman | 2019-06-21 | Build site. |
| Rmd | d11ce2b | brimittleman | 2019-06-21 | fix plots |
| html | 660979c | brimittleman | 2019-06-13 | Build site. |
| Rmd | 02b669e | brimittleman | 2019-06-13 | fix big bug |
| html | 0dbb65b | brimittleman | 2019-05-22 | Build site. |
| Rmd | d3df9c3 | brimittleman | 2019-05-22 | fix plots |
| html | 6af55a2 | brimittleman | 2019-05-21 | Build site. |
| Rmd | 2fd13bb | brimittleman | 2019-05-21 | add location plots |
| html | a88eedf | brimittleman | 2019-05-20 | Build site. |
| html | ccebe33 | brimittleman | 2019-04-24 | Build site. |
| html | 74a1372 | brimittleman | 2019-04-24 | Build site. |
| html | 012892d | brimittleman | 2019-04-24 | Build site. |
| html | 1fb7086 | brimittleman | 2019-04-23 | Build site. |
In this analysis I will create discriptive plots for the PAS identified in the 54 LCLs.
library(workflowr)This is workflowr version 1.4.0
Run ?workflowr for help getting startedlibrary(tidyverse)── Attaching packages ────────────────────────────────────────────────────────────────────────────────────── tidyverse 1.2.1 ──✔ ggplot2 3.1.1 ✔ purrr 0.3.2
✔ tibble 2.1.1 ✔ dplyr 0.8.0.1
✔ tidyr 0.8.3 ✔ stringr 1.3.1
✔ readr 1.3.1 ✔ forcats 0.3.0 ── Conflicts ───────────────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()library(reshape2)
Attaching package: 'reshape2'The following object is masked from 'package:tidyr':
smithslibrary(cowplot)
Attaching package: 'cowplot'The following object is masked from 'package:ggplot2':
ggsavePeaks per gene:
I want to plot how many genes have 0, 1, 2 and more than 2 PAS in the set. I need to join my PAS with the annotation to find out how many genes have 0 PAS.
pas=read.table("../data/PAS/APAPAS_GeneLocAnno.5perc.bed", header = F, stringsAsFactors = F, col.names = c("Chr", "start", "end", "PeakID", "score", "strand")) %>% separate(PeakID, into=c("peaknum", "geneAnno"), sep=":") %>% separate(geneAnno, into=c("Gene", "Loc"),sep="_")
pasbygene= pas %>% group_by(Gene) %>% summarise(PAS=n())
annotation=read.table("../../genome_anotation_data/refseq.ProteinCoding.bed", col.names = c("chr", "start", "end", "Gene", "score", "strand")) %>% dplyr::select(Gene) %>% unique()
PASallgene=annotation %>% left_join(pasbygene, by="Gene") %>% replace_na(list(PAS=0)) Warning: Column `Gene` joining factor and character vector, coercing into
character vector#group with 0,1,2,more than 2
PASallgene_grouped=PASallgene %>% mutate(Zero=ifelse(PAS==0,1, 0), One=ifelse(PAS==1,1,0), Multiple=ifelse(PAS>1,1,0))Plot this:
Genes=c(sum(PASallgene_grouped$Zero),sum(PASallgene_grouped$One),sum(PASallgene_grouped$Multiple))
PAS=c("Zero", "One", "Multiple")
AllPAS=c(sum(PASallgene_grouped$Zero),sum(PASallgene_grouped$One),sum(PASallgene_grouped$Multiple))
GenebyPAS=as.data.frame(cbind(PAS,AllPAS))
GenebyPAS$AllPAS=as.numeric(as.character(GenebyPAS$AllPAS))
allPASplot=ggplot(GenebyPAS, aes(x="",y=AllPAS, fill=PAS)) + geom_bar(stat="identity", width=.5) + scale_fill_brewer(palette="GnBu") + labs(title="Identified PAS per Gene", y="Genes",x="All Indentified PAS")
allPASplot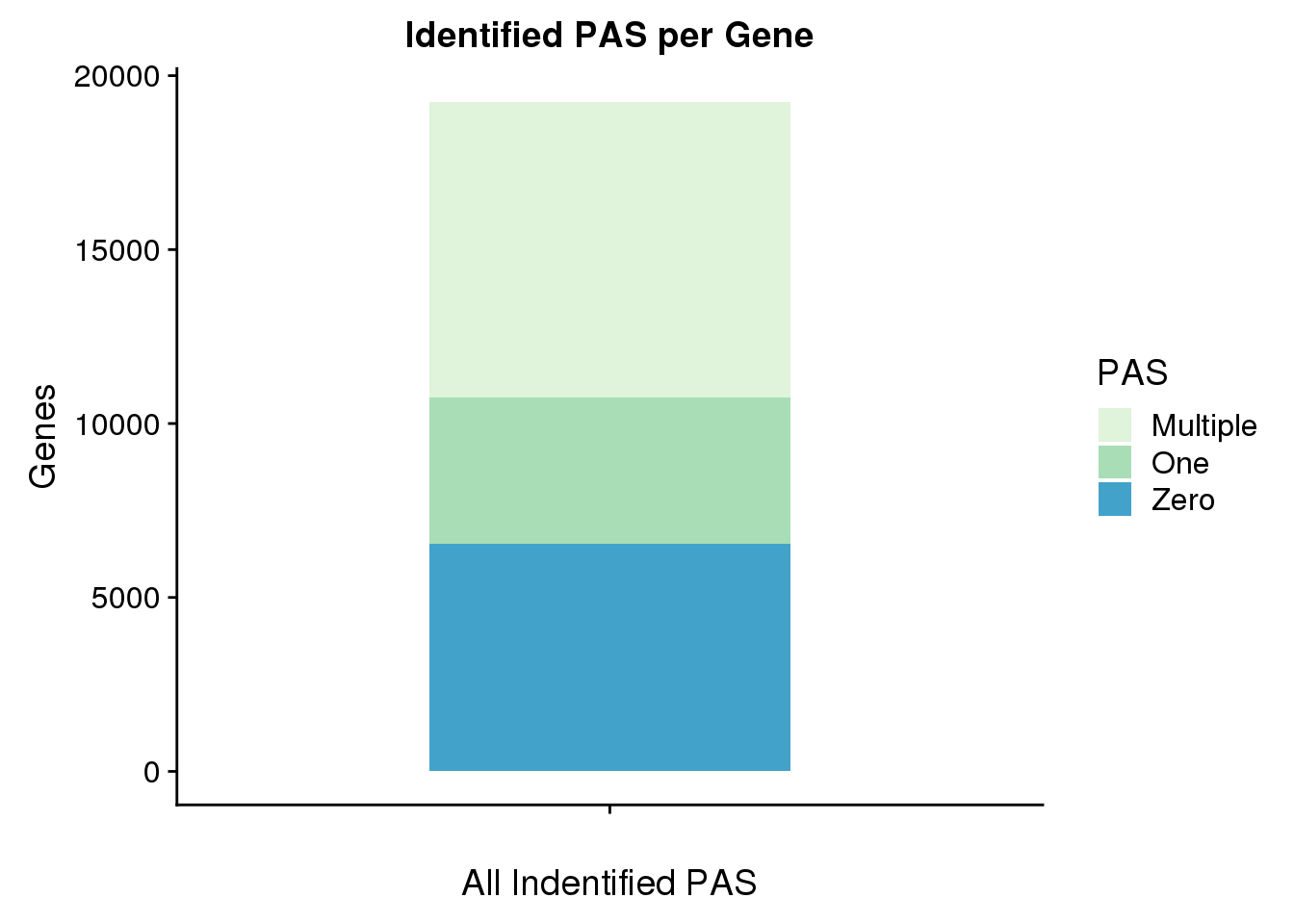
ggsave(allPASplot, file="../output/GeneswithAPApotentialAllPAS.png", width=3, height=5)Subset and get stats for UTR
pasUTR=pas %>% filter(Loc=="utr3") %>% group_by(Gene) %>% summarise(PAS=n())
pasUTR_allgene=annotation %>% full_join(pasUTR, by="Gene") %>% replace_na(list(PAS=0)) Warning: Column `Gene` joining factor and character vector, coercing into
character vectorPASUTRallgene_grouped=pasUTR_allgene %>% mutate(Zero=ifelse(PAS==0,1, 0), One=ifelse(PAS==1,1,0), Multiple=ifelse(PAS>1,1,0))
GenesUTR=c(sum(PASUTRallgene_grouped$Zero),sum(PASUTRallgene_grouped$One),sum(PASUTRallgene_grouped$Multiple))
UTR=c(sum(PASUTRallgene_grouped$Zero),sum(PASUTRallgene_grouped$One),sum(PASUTRallgene_grouped$Multiple))
GenebyPASUTR=as.data.frame(cbind(PAS,UTR))
GenebyPASUTR$UTR=as.numeric(as.character(GenebyPASUTR$UTR))
ggplot(GenebyPASUTR, aes(x="",y=UTR, fill=PAS)) + geom_bar(stat="identity")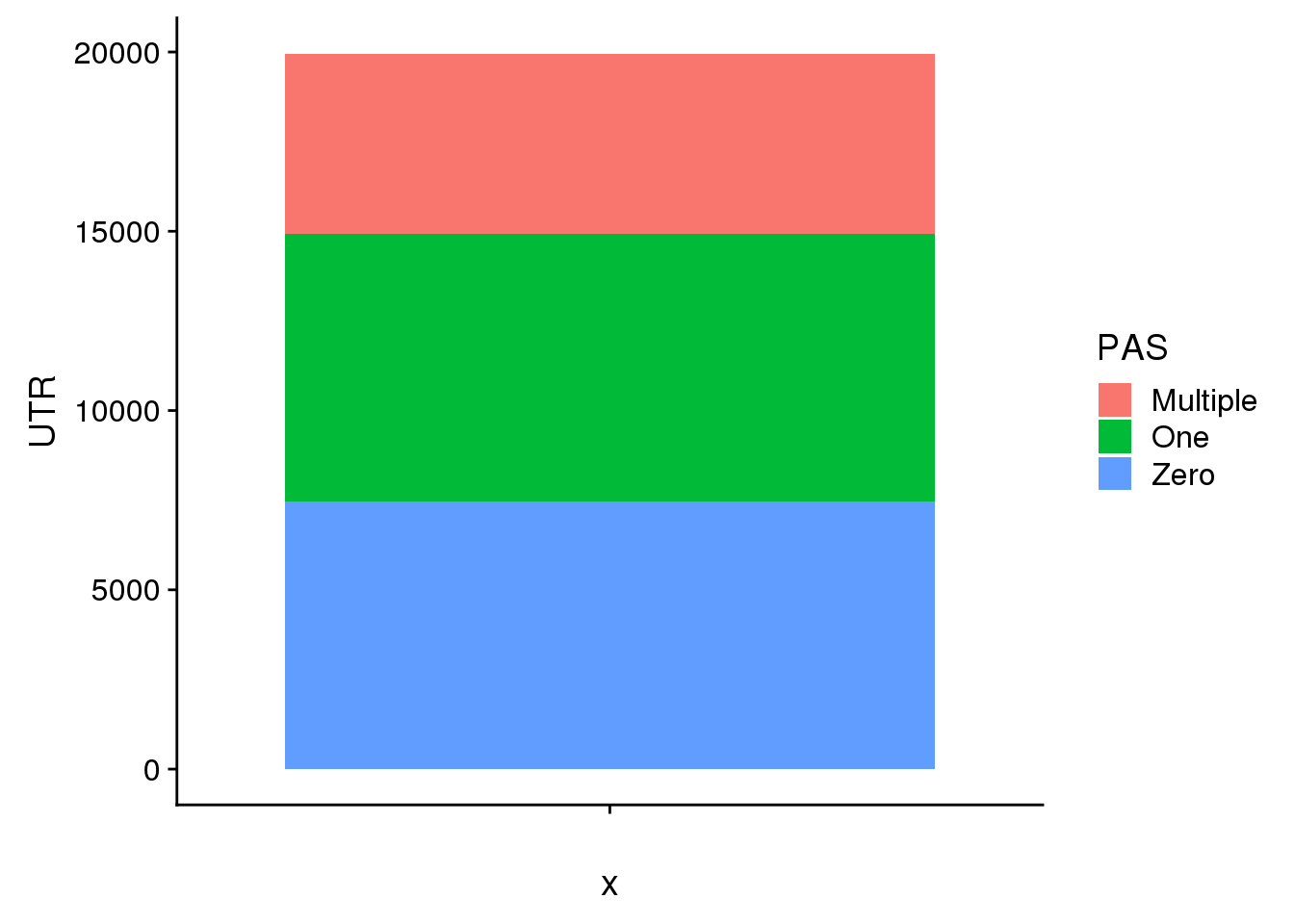
Subset and get stats for Intron
pasIntron=pas %>% filter(Loc=="intron" | Loc=='utr3') %>% group_by(Gene) %>% summarise(PAS=n())
pasIntron_allgene=annotation %>% full_join(pasIntron, by="Gene") %>% replace_na(list(PAS=0)) Warning: Column `Gene` joining factor and character vector, coercing into
character vectorpasIntronallgene_grouped=pasIntron_allgene %>% mutate(Zero=ifelse(PAS==0,1, 0), One=ifelse(PAS==1,1,0), Multiple=ifelse(PAS>1,1,0))
UTRandIntron=c(sum(pasIntronallgene_grouped$Zero),sum(pasIntronallgene_grouped$One),sum(pasIntronallgene_grouped$Multiple))
GenebyPASIntron=as.data.frame(cbind(PAS,UTRandIntron))
GenebyPASIntron$UTRandIntron=as.numeric(as.character(GenebyPASIntron$UTRandIntron))
ggplot(GenebyPASIntron, aes(x="",y=UTRandIntron, fill=PAS)) + geom_bar(stat="identity")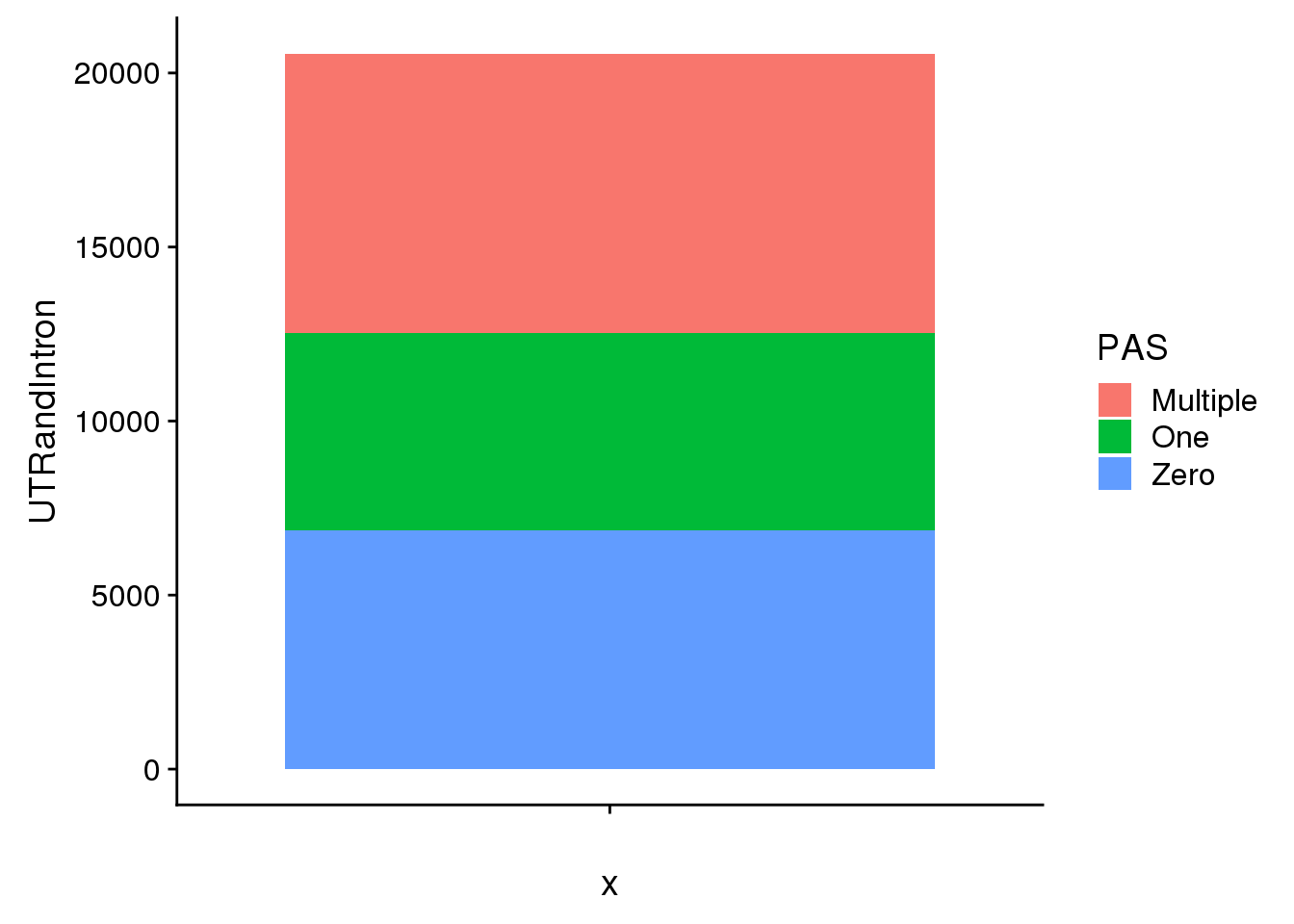
Make these side by side:
GenebyPASUTR_melt=melt(GenebyPASUTR, id.vars = "PAS", value.name = "Genes", variable.name = "Set")
GenebyPAS_melt=melt(GenebyPAS, id.vars = "PAS", value.name = "Genes", variable.name = "Set")
GenebyPASIntron_melt=melt(GenebyPASIntron, id.vars = "PAS", value.name = "Genes", variable.name = "Set")
GenebyPAStoplot=rbind(GenebyPAS_melt,GenebyPASUTR_melt,GenebyPASIntron_melt)
geneswithAPA=ggplot(GenebyPAStoplot, aes(x=Set,y=Genes, fill=PAS, by=Set)) + geom_bar(stat="identity")+ scale_fill_brewer(palette="YlGnBu") + labs(title="Genes with APA poential")
geneswithAPA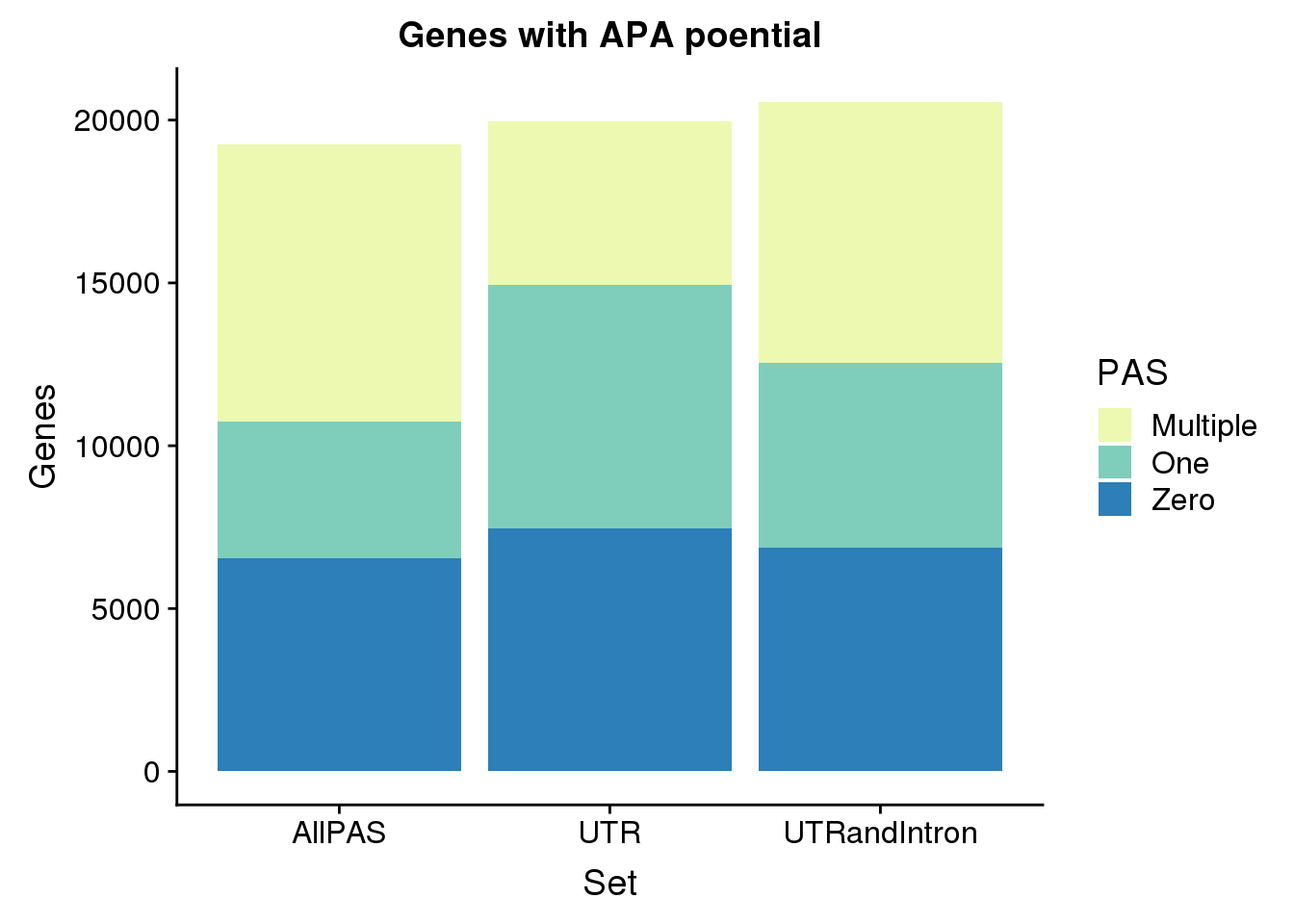
ggsave(geneswithAPA, file="../output/GeneswithAPApotential.png")Saving 7 x 5 in imageGenebyPAStoplot PAS Set Genes
1 Zero AllPAS 6550
2 One AllPAS 4198
3 Multiple AllPAS 8495
4 Zero UTR 7463
5 One UTR 7466
6 Multiple UTR 5024
7 Zero UTRandIntron 6868
8 One UTRandIntron 5672
9 Multiple UTRandIntron 8015Location of PAS
PAS_loc=pas %>% group_by(Loc) %>% summarise(nPAS=n())
loclabel=c("Coding", "Downstream", "Intronic", "3' UTR", "5' UTR")
PASLocPlot=ggplot(PAS_loc, aes(x=Loc, y=nPAS, fill=Loc)) + geom_bar(stat="identity",width=.5)+ scale_fill_brewer(palette = "YlGnBu") + labs(x="Gene location", y="Number of identified PAS", title="Location distribution for identified PAS") + theme(legend.position = "none")+ scale_x_discrete(labels= loclabel)+theme(axis.text.x = element_text(angle = 90, hjust = 1))
PASLocPlot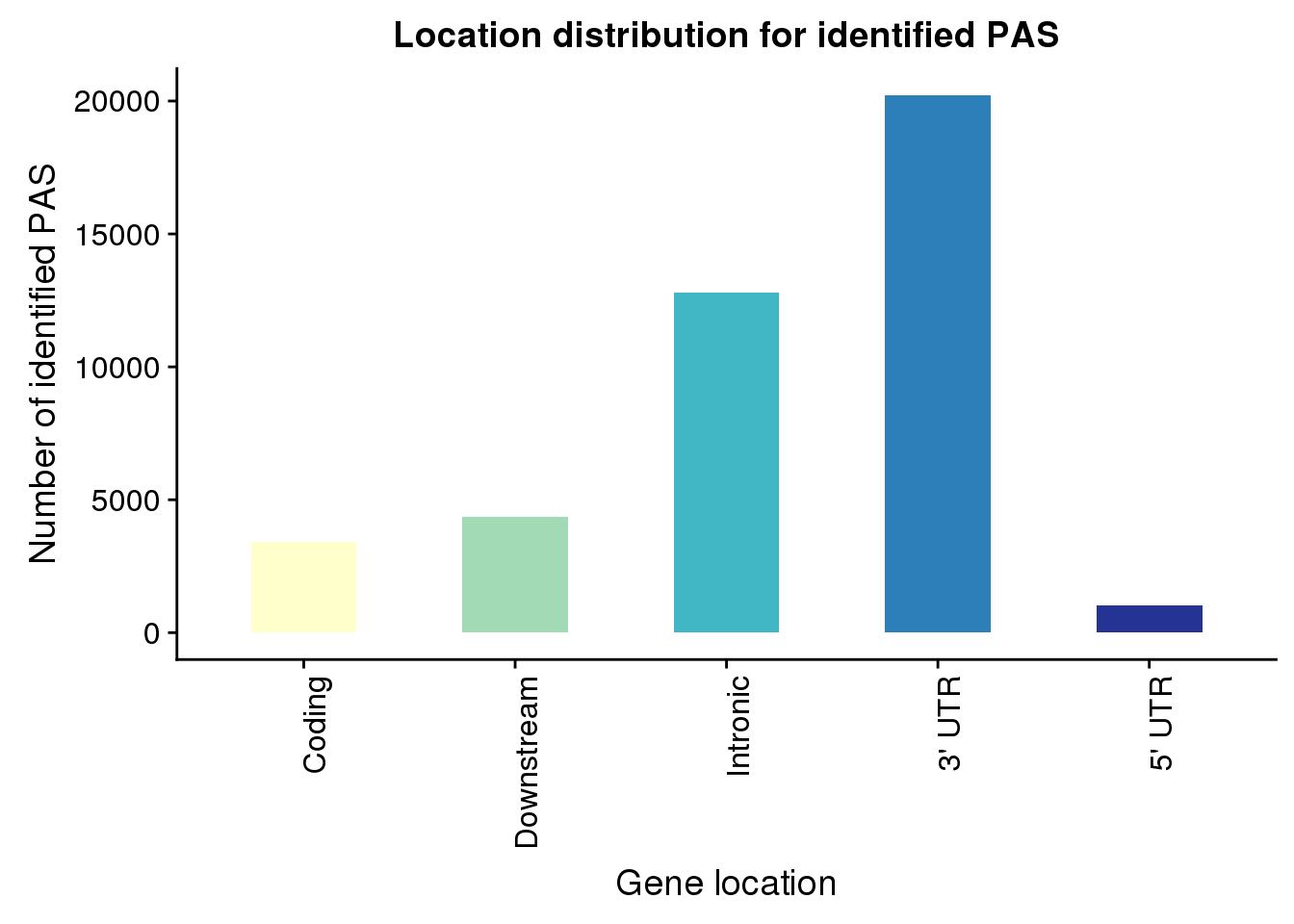
ggsave(PASLocPlot, file="../output/PASlocation.png")Saving 7 x 5 in imageNumber of genes with apa by cutoff
I want to make a script that takes a cuttoff and tells me how many gens have 0,1, >1 PAS. This way I can put these together to make stacked barplots:
I can make the plot for total then again for nuclear.
The annotaiton is annotation:
totapaanno=read.table("../data/phenotype/APApeak_Phenotype_GeneLocAnno.Total.fc",header = T,stringsAsFactors = F)
indiv=colnames(totapaanno)[2:53]
totapanum=read.table("../data/phenotype/APApeak_Phenotype_GeneLocAnno.Total.CountsOnlyNumeric",header = F, col.names = indiv)
totapa_mean=rowMeans(totapanum)
totapaMeananno=as.data.frame(cbind(ID=totapaanno$chrom, meanUsage=totapa_mean))
totapaMeananno$meanUsage=as.numeric(as.character(totapaMeananno$meanUsage))
totapaMeananno$ID=as.character(totapaMeananno$ID)
write.table(totapaMeananno, file="../data/PAS/TotalPASMeanUsage.txt", col.names = T, row.names = F, quote=F,sep="\t")genesbycuttoff_tot=function(fraction){
totapaMeananno_filt=totapaMeananno %>% filter(meanUsage >=fraction) %>% separate(ID, into=c("chrom", "start","end", "peakID"),sep=":") %>% separate(peakID, into=c("Gene","loc", "strand", "peak"),sep="_") %>% group_by(Gene) %>% summarise(PAS=n())
PASallgene=annotation %>% full_join(totapaMeananno_filt, by="Gene") %>% replace_na(list(PAS=0))
PASallgene_cat=PASallgene %>% mutate(Category=ifelse(PAS==0,"Zero", ifelse(PAS==1, "One", "Multiple"))) %>% group_by(Category) %>% summarise(NPer=n())
return(PASallgene_cat$NPer)
}#multiple, one, zero
categories=c("Multiple_PAS", "One_PAS", "Zero_PAS")
FullDF=as.data.frame(cbind(categories))
cutoffs=seq(from=0, to=.5, by=.05)
for (val in cutoffs[1:10])
{
FullDF=cbind(FullDF,val=genesbycuttoff_tot(val))
}Warning: Expected 4 pieces. Additional pieces discarded in 3 rows [12630,
12631, 12632].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 3 rows [4597,
4598, 4599].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 3 rows [3413,
3414, 3415].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 2 rows [2856,
2857].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 2 rows [2508,
2509].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 1 rows [2223].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 1 rows [1985].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 1 rows [1789].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 1 rows [1641].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 1 rows [1520].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorcolnames(FullDF)=c("Category",cutoffs[1:10])Melt:
fullDF_melt=melt(FullDF,id.vars = "Category",variable.name = "Cutoff", value.name = "NGenes") %>% mutate(propGene=NGenes/nrow(annotation))
totalPropgenes=ggplot(fullDF_melt,aes(x=Cutoff, y=propGene, by=Category, fill=Category)) + geom_bar(width=1, stat="identity") + labs(title="Total Fraction", y="Proportion of 19,243 genes", x="Usage Filter cutoff") + theme(axis.text.x = element_text(angle = 90, hjust =1)) + scale_x_discrete(name="Usage Filter cutoff", breaks=c("0","0.1","0.2", "0.3", "0.4","0.5"))+ theme(text = element_text(size=16),legend.position = "bottom")+scale_fill_brewer(name = "", labels = c("Multiple PAS", "One PAS", "Zero Identified PAS"),palette="Dark2")
totalPropgenes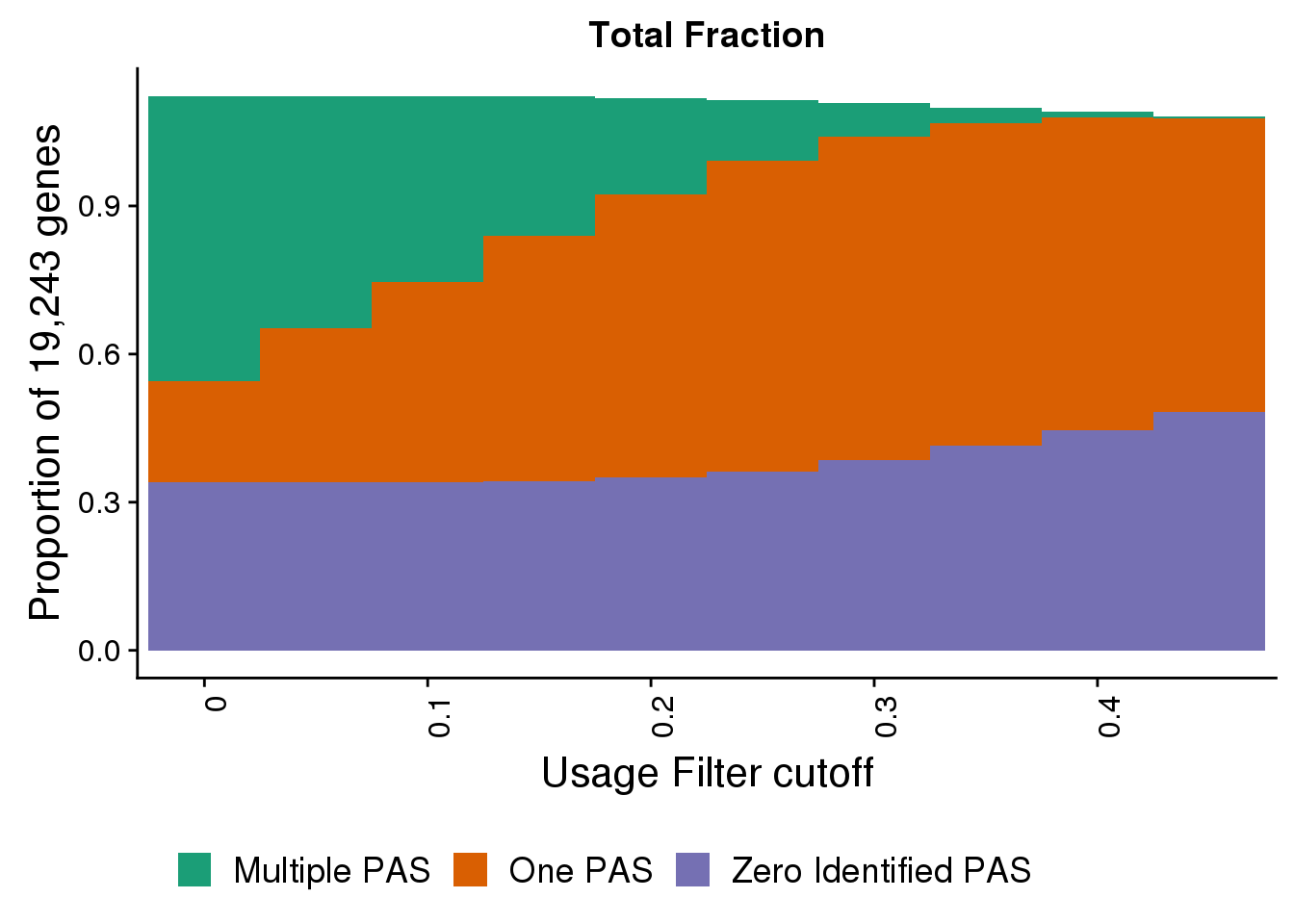
Nuclear:
nucapaanno=read.table("../data/phenotype/APApeak_Phenotype_GeneLocAnno.Nuclear.fc",header = T,stringsAsFactors = F)
nucapanum=read.table("../data/phenotype/APApeak_Phenotype_GeneLocAnno.Nuclear.CountsOnlyNumeric",header = F, col.names = indiv)
nucapa_mean=rowMeans(nucapanum)
nucapaMeananno=as.data.frame(cbind(ID=nucapaanno$chrom, meanUsage=nucapa_mean))
nucapaMeananno$meanUsage=as.numeric(as.character(nucapaMeananno$meanUsage))
nucapaMeananno$ID=as.character(nucapaMeananno$ID)
write.table(nucapaMeananno, file="../data/PAS/NuclearPASMeanUsage.txt", col.names = T, row.names = F, quote=F,sep="\t")genesbycuttoff_nuc=function(fraction){
nucapaMeananno_filt=nucapaMeananno %>% filter(meanUsage >=fraction) %>% separate(ID, into=c("chrom", "start","end", "peakID"),sep=":") %>% separate(peakID, into=c("Gene","loc", "strand", "peak"),sep="_") %>% group_by(Gene) %>% summarise(PAS=n())
PASallgene=annotation %>% full_join(nucapaMeananno_filt, by="Gene") %>% replace_na(list(PAS=0))
PASallgene_cat=PASallgene %>% mutate(Category=ifelse(PAS==0,"Zero", ifelse(PAS==1, "One", "Multiple"))) %>% group_by(Category) %>% summarise(NPer=n())
return(PASallgene_cat$NPer)
}#multiple, one, zero
FullDFNuc=as.data.frame(cbind(categories))
for (val in cutoffs[1:10])
{
FullDFNuc=cbind(FullDFNuc,val=genesbycuttoff_nuc(val))
}Warning: Expected 4 pieces. Additional pieces discarded in 3 rows [12630,
12631, 12632].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 3 rows [5138,
5139, 5140].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 3 rows [3674,
3675, 3676].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 3 rows [2980,
2981, 2982].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 2 rows [2546,
2547].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 1 rows [2259].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 1 rows [2012].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 1 rows [1804].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 1 rows [1640].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorWarning: Expected 4 pieces. Additional pieces discarded in 1 rows [1500].Warning: Column `Gene` joining factor and character vector, coercing into
character vectorcolnames(FullDFNuc)=c("Category",cutoffs[1:10])FullDFNuc_melt=melt(FullDFNuc,id.vars = "Category",variable.name = "Cutoff", value.name = "NGenes") %>% mutate(propGene=NGenes/nrow(annotation))
nuclearPropgenes=ggplot(FullDFNuc_melt,aes(x=Cutoff, y=propGene, by=Category, fill=Category)) + geom_bar( width=1,stat="identity") + labs(title="Nuclear Fraction",y="Proportion of 19,243 genes", x="Usage Filter cutoff") + scale_x_discrete(name="Usage Filter cutoff", breaks=c("0","0.1","0.2", "0.3", "0.4","0.5")) + theme(text = element_text(size=16), legend.position = "bottom")+ scale_fill_brewer(name = "", labels = c("Multiple PAS", "One PAS", "Zero Identified PAS"),palette="Dark2")
nuclearPropgenes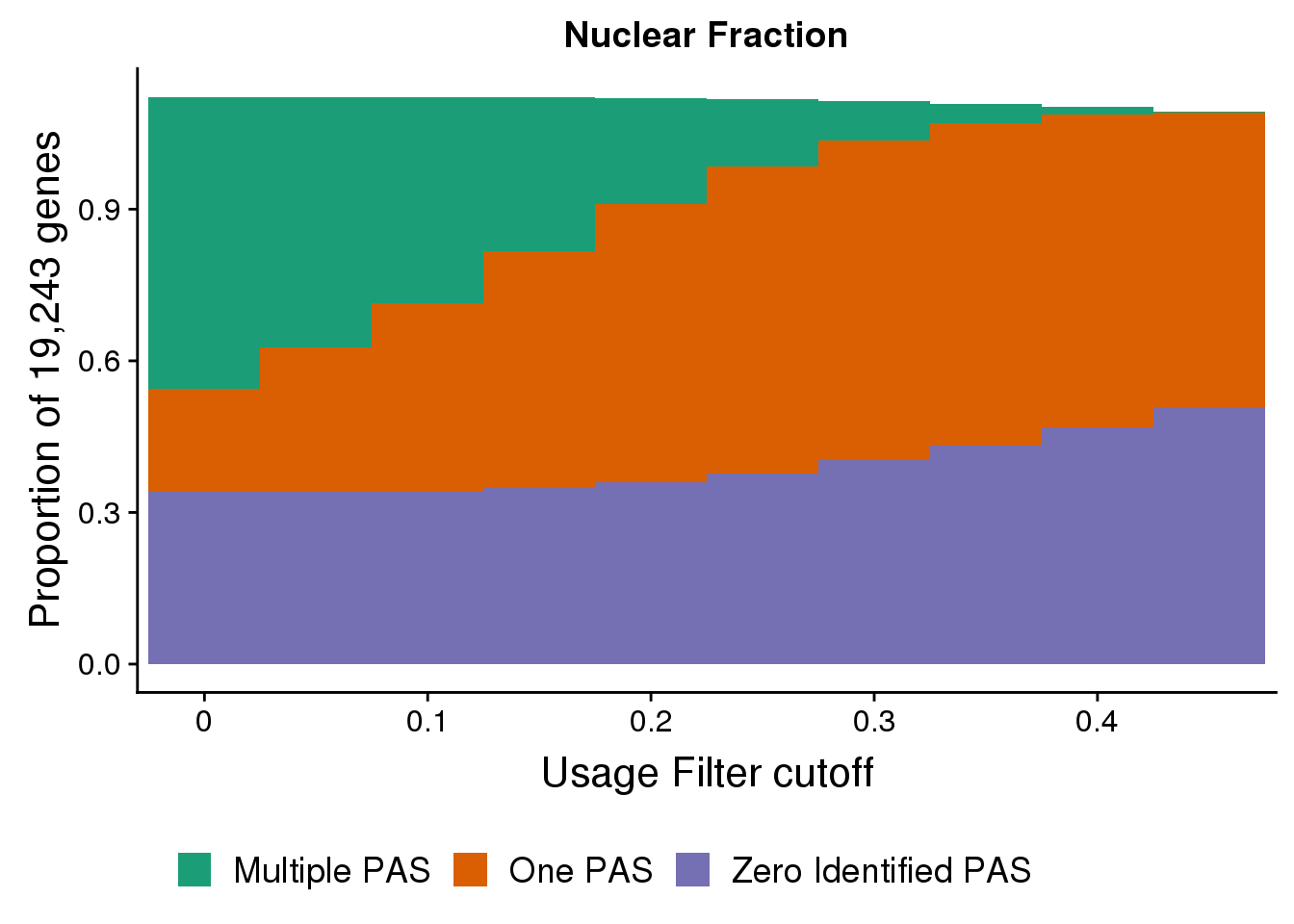
title_theme <- ggdraw() +
draw_label("Proportion of Genes by PAS identification", x = 0.28, hjust = 0,size=20, fontface="bold")
withouttitle=plot_grid(totalPropgenes,nuclearPropgenes, rel_widths = c(1, 1))
plot_grid(title_theme,withouttitle,ncol = 1, rel_heights = c(0.2, 1))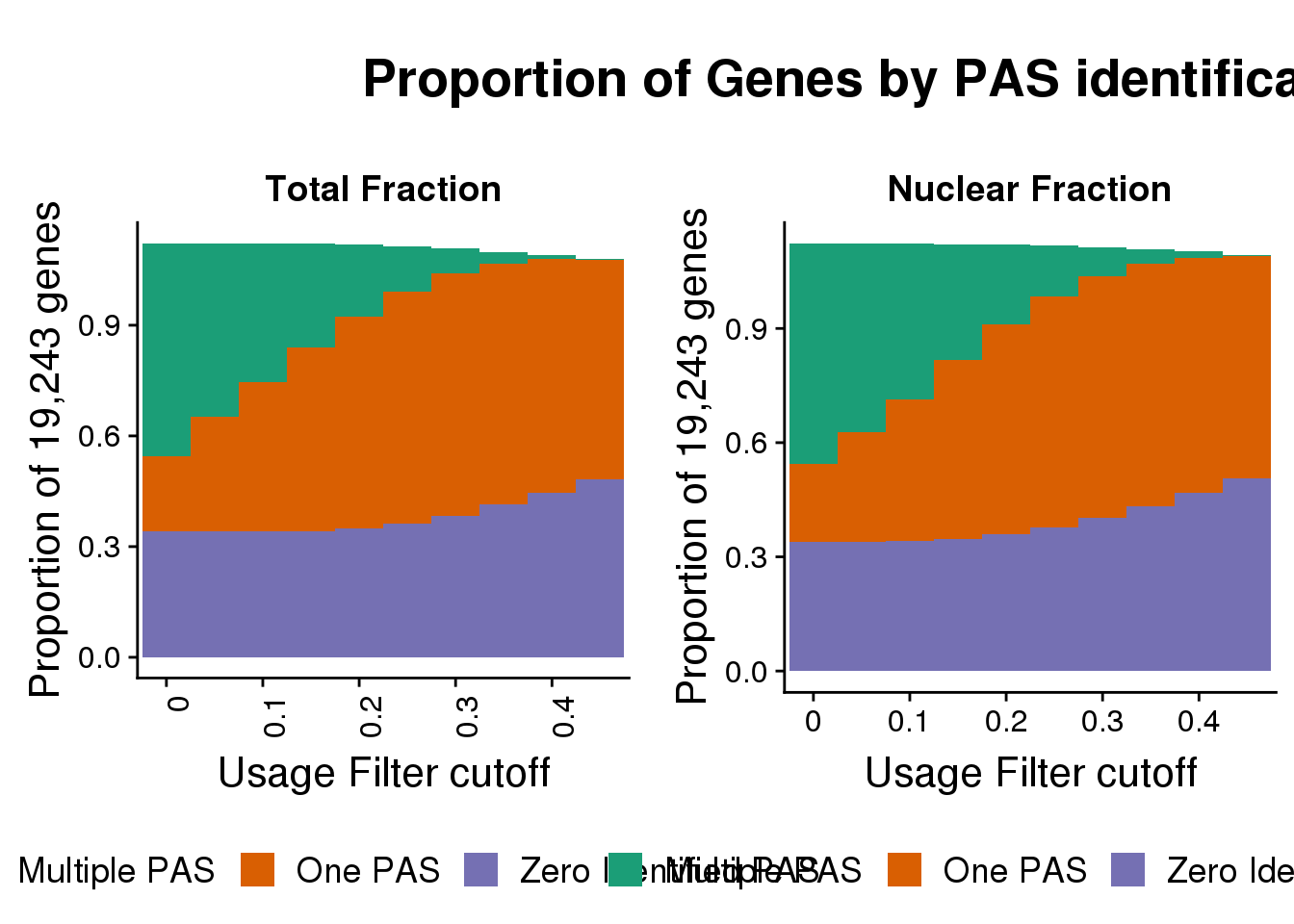
Location of PAS by filter
I will make these plots but the categories will be location of the PAS.
locbycutoff_tot=function(fraction){
totapaMeananno_filt=totapaMeananno %>% filter(meanUsage >=fraction) %>% separate(ID, into=c("chrom", "start","end", "peakID"),sep=":") %>% separate(peakID, into=c("Gene","loc", "strand", "peak"),sep="_") %>% group_by(loc) %>% summarise(PerLoc=n()) %>%filter(loc!= "008559")
return(totapaMeananno_filt$PerLoc)
}locations=c("cds", "end", "intron", "utr3", "utr5")
FullDF_loc=as.data.frame(cbind(locations))
cutoffs=seq(from=0, to=.5, by=.05)
for (val in cutoffs[1:10])
{
FullDF_loc=cbind(FullDF_loc,val=locbycutoff_tot(val))
}Warning: Expected 4 pieces. Additional pieces discarded in 3 rows [12630,
12631, 12632].Warning: Expected 4 pieces. Additional pieces discarded in 3 rows [4597,
4598, 4599].Warning: Expected 4 pieces. Additional pieces discarded in 3 rows [3413,
3414, 3415].Warning: Expected 4 pieces. Additional pieces discarded in 2 rows [2856,
2857].Warning: Expected 4 pieces. Additional pieces discarded in 2 rows [2508,
2509].Warning: Expected 4 pieces. Additional pieces discarded in 1 rows [2223].Warning: Expected 4 pieces. Additional pieces discarded in 1 rows [1985].Warning: Expected 4 pieces. Additional pieces discarded in 1 rows [1789].Warning: Expected 4 pieces. Additional pieces discarded in 1 rows [1641].Warning: Expected 4 pieces. Additional pieces discarded in 1 rows [1520].colnames(FullDF_loc)=c("Location",cutoffs[1:10])Melt:
FullDF_loc_melt=melt(FullDF_loc,id.vars = "Location",variable.name = "Cutoff", value.name = "NPas") %>% group_by(Cutoff) %>% mutate(propPAS=NPas/sum(NPas))
totplotloc=ggplot(FullDF_loc_melt,aes(x=Cutoff, y=propPAS, by=Location, fill=Location)) + geom_bar(width=1, stat="identity") + labs(title="Total Fraction",y="Proportion of PAS", x="Usage Filter cutoff")+ theme(axis.text.x = element_text(angle = 90, hjust = 1)) + scale_x_discrete(name="Usage Filter cutoff", breaks=c("0","0.1","0.2", "0.3", "0.4","0.5"))+ theme(text = element_text(size=16), legend.position = "bottom")+ scale_fill_brewer(name = "", labels = c("Coding", "5KB downstread", "Intronic", "3' UTR", "5' UTR"),palette="Dark2")+guides(fill=guide_legend(nrow=2,byrow=TRUE))Nuclear
locbycutoff_nuc=function(fraction){
nucapaMeananno_filt=nucapaMeananno %>% filter(meanUsage >=fraction) %>% separate(ID, into=c("chrom", "start","end", "peakID"),sep=":") %>% separate(peakID, into=c("Gene","loc", "strand", "peak"),sep="_") %>% group_by(loc) %>% summarise(PerLoc=n()) %>%filter(loc!= "008559")
return(nucapaMeananno_filt$PerLoc)
}NucFullDF_loc=as.data.frame(cbind(locations))
cutoffs=seq(from=0, to=.5, by=.05)
for (val in cutoffs)
{
NucFullDF_loc=cbind(NucFullDF_loc,val=locbycutoff_nuc(val))
}Warning: Expected 4 pieces. Additional pieces discarded in 3 rows [12630,
12631, 12632].Warning: Expected 4 pieces. Additional pieces discarded in 3 rows [5138,
5139, 5140].Warning: Expected 4 pieces. Additional pieces discarded in 3 rows [3674,
3675, 3676].Warning: Expected 4 pieces. Additional pieces discarded in 3 rows [2980,
2981, 2982].Warning: Expected 4 pieces. Additional pieces discarded in 2 rows [2546,
2547].Warning: Expected 4 pieces. Additional pieces discarded in 1 rows [2259].Warning: Expected 4 pieces. Additional pieces discarded in 1 rows [2012].Warning: Expected 4 pieces. Additional pieces discarded in 1 rows [1804].Warning: Expected 4 pieces. Additional pieces discarded in 1 rows [1640].Warning: Expected 4 pieces. Additional pieces discarded in 1 rows [1500].Warning: Expected 4 pieces. Additional pieces discarded in 1 rows [1359].colnames(NucFullDF_loc)=c("Location",cutoffs)Melt:
NucFullDF_locMelt=melt(NucFullDF_loc,id.vars = "Location",variable.name = "Cutoff", value.name = "NPas") %>% group_by(Cutoff) %>% mutate(propPAS=NPas/sum(NPas))
nucplotloc=ggplot(NucFullDF_locMelt,aes(x=Cutoff, y=propPAS, by=Location, fill=Location)) + geom_bar(width = 1, stat="identity") + labs(title="Nuclear Fraction",y="Proportion of PAS", x="Usage Filter cutoff")+ theme(axis.text.x = element_text(angle = 90, hjust = 1)) + scale_x_discrete(name="Usage Filter cutoff", breaks=c("0","0.1","0.2", "0.3", "0.4","0.5"))+ theme(text = element_text(size=16), legend.position = "bottom", legend.box="horizontal")+ scale_fill_brewer(name = "", labels = c("Coding", "5KB downstread", "Intronic", "3' UTR", "5' UTR"),palette="Dark2") + guides(fill=guide_legend(nrow=2,byrow=TRUE))
nucplotloc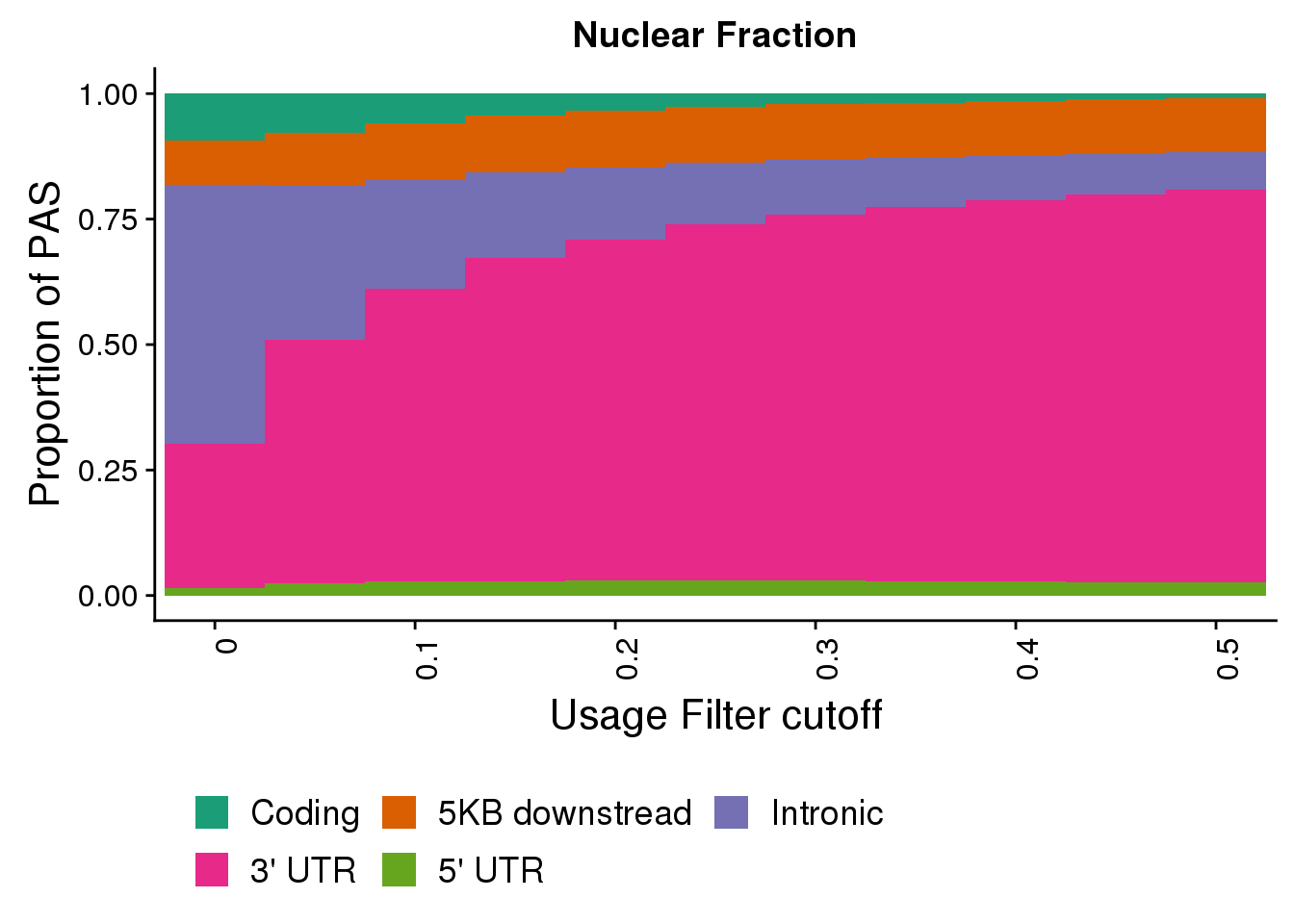
Plot next to eachother
title_loc <- ggdraw() +
draw_label("Location of Identified PAS", x = 0.35, hjust = 0,size=20, fontface="bold")
locPlots=plot_grid(totplotloc,nucplotloc)
locPlotswtitle=plot_grid(title_loc,locPlots,ncol = 1, rel_heights = c(0.2, 1))locPlotswtitle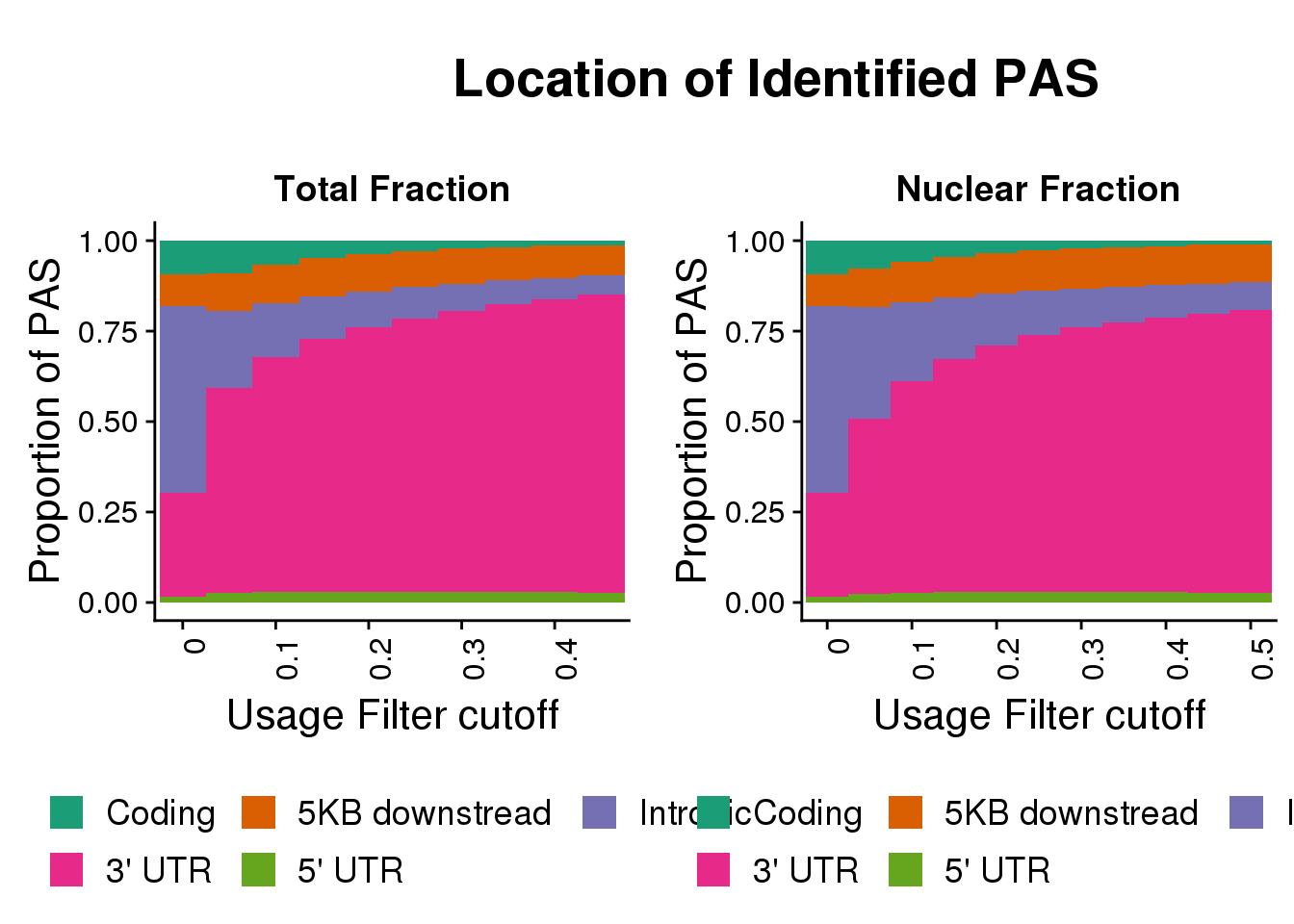
Nuclear specific PAS (only identified at 5% in nuclear)
NucPAS=read.table("../data/PAS/NuclearPASMeanUsage.txt",header =T,stringsAsFactors = F ) %>% filter(meanUsage > 0.05)
TotPAS=read.table("../data/PAS/TotalPASMeanUsage.txt",header=T, stringsAsFactors = F) %>% filter(meanUsage>0.05)
nucspecific=NucPAS %>% anti_join(TotPAS, by="ID")
totspecific=TotPAS %>% anti_join(NucPAS, by="ID")nucspecific_sep=nucspecific %>% separate(ID, into=c("chr", "start", "end", "geneID"), sep=":") %>% separate(geneID, into=c("gene","loc", "strand","pas"),sep="_")
ggplot(nucspecific_sep, aes(x=loc)) +geom_bar(stat="count")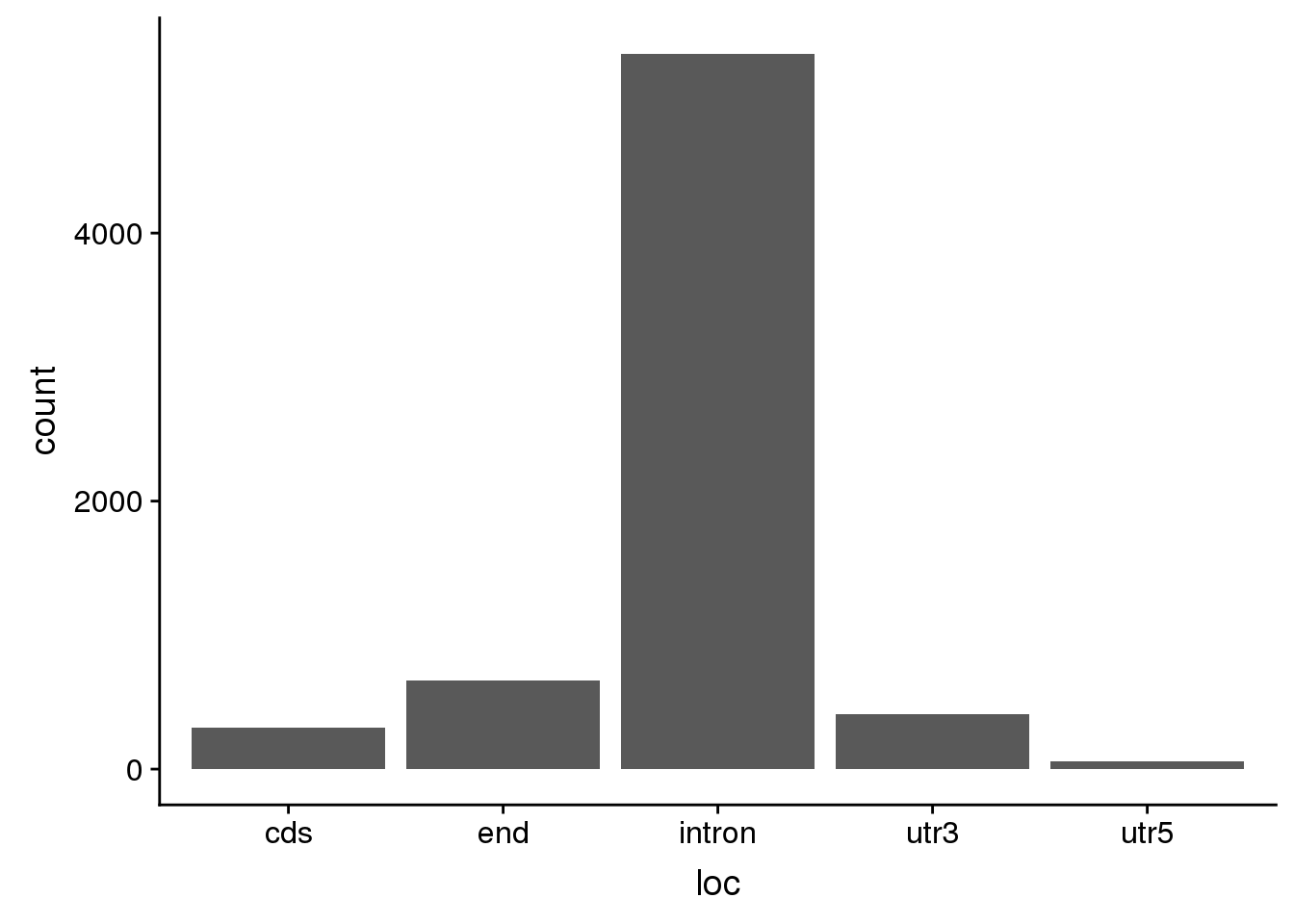
| Version | Author | Date |
|---|---|---|
| 1419181 | brimittleman | 2019-08-01 |
summary(nucspecific_sep$meanUsage) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.05019 0.05788 0.06942 0.08064 0.09135 0.42500 nucspecific_sep %>% group_by(loc) %>% summarise(nLoc=n())# A tibble: 5 x 2
loc nLoc
<chr> <int>
1 cds 308
2 end 659
3 intron 5339
4 utr3 409
5 utr5 56nucspecific_sep_int=nucspecific_sep %>% filter(loc=="intron")
summary(nucspecific_sep_int$meanUsage) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.05019 0.05885 0.07096 0.08190 0.09288 0.38154 totspecific_sep=totspecific %>% separate(ID, into=c("chr", "start", "end", "geneID"), sep=":") %>% separate(geneID, into=c("gene","loc", "strand","pas"),sep="_")
ggplot(totspecific_sep, aes(x=loc)) +geom_bar(stat="count")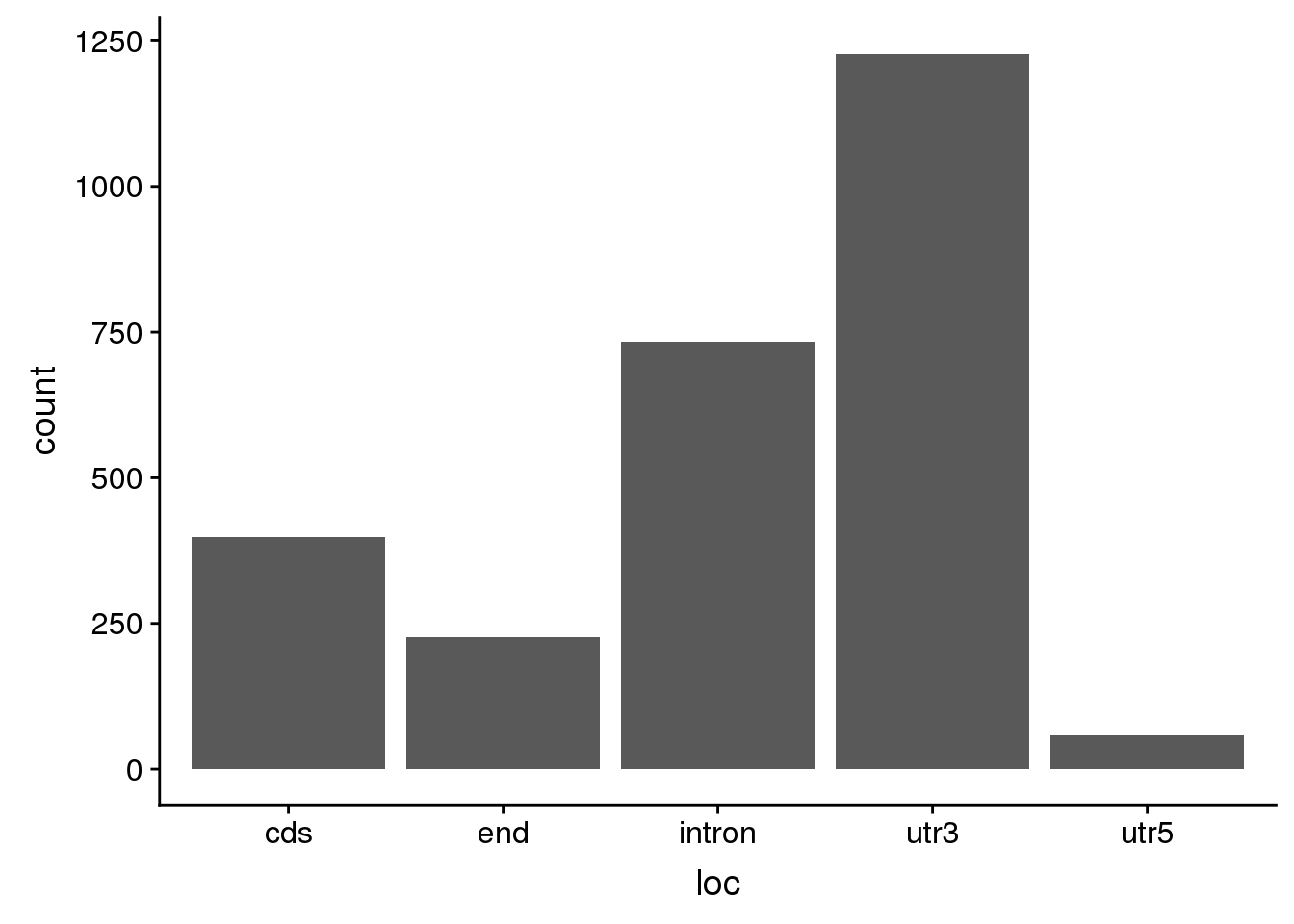
| Version | Author | Date |
|---|---|---|
| 1419181 | brimittleman | 2019-08-01 |
summary(totspecific_sep$meanUsage) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.05019 0.05577 0.06385 0.07341 0.07976 0.30769
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_0.9.4 reshape2_1.4.3 forcats_0.3.0 stringr_1.3.1
[5] dplyr_0.8.0.1 purrr_0.3.2 readr_1.3.1 tidyr_0.8.3
[9] tibble_2.1.1 ggplot2_3.1.1 tidyverse_1.2.1 workflowr_1.4.0
loaded via a namespace (and not attached):
[1] tidyselect_0.2.5 haven_1.1.2 lattice_0.20-38
[4] colorspace_1.3-2 generics_0.0.2 htmltools_0.3.6
[7] yaml_2.2.0 utf8_1.1.4 rlang_0.4.0
[10] pillar_1.3.1 glue_1.3.0 withr_2.1.2
[13] RColorBrewer_1.1-2 modelr_0.1.2 readxl_1.1.0
[16] plyr_1.8.4 munsell_0.5.0 gtable_0.2.0
[19] cellranger_1.1.0 rvest_0.3.2 evaluate_0.12
[22] labeling_0.3 knitr_1.20 fansi_0.4.0
[25] highr_0.7 broom_0.5.1 Rcpp_1.0.0
[28] scales_1.0.0 backports_1.1.2 jsonlite_1.6
[31] fs_1.3.1 hms_0.4.2 digest_0.6.18
[34] stringi_1.2.4 grid_3.5.1 rprojroot_1.3-2
[37] cli_1.1.0 tools_3.5.1 magrittr_1.5
[40] lazyeval_0.2.1 crayon_1.3.4 whisker_0.3-2
[43] pkgconfig_2.0.2 xml2_1.2.0 lubridate_1.7.4
[46] assertthat_0.2.0 rmarkdown_1.10 httr_1.3.1
[49] rstudioapi_0.10 R6_2.3.0 nlme_3.1-137
[52] git2r_0.25.2 compiler_3.5.1