liftover_pipeline
Briana Mittleman
8/1/2018
Last updated: 2018-08-28
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20180801)The command
set.seed(20180801)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: f670cfa
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .RData Ignored: .Rhistory Ignored: .Rproj.user/ Untracked files: Untracked: com_threeprime.Rproj Untracked: data/PeakPerExon/ Untracked: data/PeakPerGene/ Untracked: data/comp.pheno.data.csv Untracked: data/dist_TES/ Untracked: data/dist_upexon/ Untracked: data/liftover/ Untracked: data/map.stats.csv Untracked: data/map.stats.xlsx Untracked: data/orthoPeak_quant/ Unstaged changes: Deleted: comparitive_threeprime.Rproj
Expand here to see past versions:
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | f670cfa | brimittleman | 2018-08-28 | add plot for num conserved peaks |
| html | bb880d8 | brimittleman | 2018-08-24 | Build site. |
| Rmd | c39776d | brimittleman | 2018-08-24 | add corresponding peak names |
| html | d0d4599 | brimittleman | 2018-08-21 | Build site. |
| Rmd | 843d7d1 | brimittleman | 2018-08-21 | update based on knitr profile |
| html | 7bdbd48 | brimittleman | 2018-08-17 | Build site. |
| Rmd | 392aed9 | brimittleman | 2018-08-17 | lift code and add to index |
I will use this analysis to create a pipeline I can use to liftover the peaks once I get them from the human and chimp three prime seq data.
Download files
Tool to add to conda environment:
- ucsc-liftover
Chain file from UCSC:
/project2/gilad/briana/genome_anotation_data/comp_genomes/liftover/hg38ToPanTro5.over.chain.gz
/project2/gilad/briana/genome_anotation_data/comp_genomes/liftover/panTro5ToHg38.over.chain.gz
Prepare the bed files
I want the bed files with the peaks to be in the folowing format:
chr# start end species_peakname
Resulting bed files will go in: /project2/gilad/briana/comparitive_threeprime/data/liftover
To go from the peak bed file created in the callPeaksbySpecies analysis I need to cut the file to the first four columns and add the species name to the peak.
awk '{print $1 "\t" $2 "\t" $3 "\t" "human_"$4}' /project2/gilad/briana/comparitive_threeprime/human/data/mergedPeaks_comb/filtered_APApeaks_merged_allchrom_named_human.bed > /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_humanPeaks.bedawk '{print $1 "\t" $2 "\t" $3 "\t" "chimp_"$4}' /project2/gilad/briana/comparitive_threeprime/chimp/data/mergedPeaks_comb/filtered_APApeaks_merged_allchrom_named_chimp.bed > /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_chimpPeaks.bedRun liftover
Run liftOver with ‘liftOver input.bed hg18ToHg19.over.chain.gz output.bed unlifted.bed’ I want to run it both direction. I will then lift back.
LiftForward.sh
#!/bin/bash
#SBATCH --job-name=LiftForward
#SBATCH --account=pi-yangili1
#SBATCH --time=24:00:00
#SBATCH --output=LiftForward.out
#SBATCH --error=LiftForward.err
#SBATCH --partition=broadwl
#SBATCH --mem=16G
#SBATCH --mail-type=END
module load Anaconda3
source activate comp_threeprime_env
#human to chimp
liftOver /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_humanPeaks.bed /project2/gilad/briana/genome_anotation_data/comp_genomes/liftover/hg38ToPanTro5.over.chain.gz /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_humanPeakslifted.bed /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_humanPeaksunlifted.bed
#chimp to human
liftOver /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_chimpPeaks.bed /project2/gilad/briana/genome_anotation_data/comp_genomes/liftover/panTro5ToHg38.over.chain.gz /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_chimpPeaks.lifted.bed /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_chimpPeaks.unlifted.bedLiftReverse.sh
Now the lifted human peaks are on chimp cordinates and vise-versa. I will lift back over.
#!/bin/bash
#SBATCH --job-name=LiftReverse
#SBATCH --time=24:00:00
#SBATCH --output=LiftReverse.out
#SBATCH --error=LiftReverse.err
#SBATCH --partition=broadwl
#SBATCH --mem=16G
#SBATCH --mail-type=END
module load Anaconda3
source activate comp_threeprime_env
#human to chimp back to human
liftOver /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_humanPeakslifted.bed /project2/gilad/briana/genome_anotation_data/comp_genomes/liftover/panTro5ToHg38.over.chain.gz /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_humanPeakslifted_reverse.bed /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_humanPeaksunlifted.reverse.bed
#chimp to human back to chimp
liftOver /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_chimpPeaks.lifted.bed /project2/gilad/briana/genome_anotation_data/comp_genomes/liftover/hg38ToPanTro5.over.chain.gz /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_chimpPeaks.lifted.reverse.bed /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_chimpPeaks.unlifted.reverse.bed
LiftFinal.sh
I now have lifted back and I want to go forward one more time to get the final list i should use.
#!/bin/bash
#SBATCH --job-name=LiftFinal
#SBATCH --time=24:00:00
#SBATCH --output=LiftFinal.out
#SBATCH --error=LiftFinal.err
#SBATCH --partition=broadwl
#SBATCH --mem=16G
#SBATCH --mail-type=END
module load Anaconda3
source activate comp_threeprime_env
#human to chimp back to human - final lift to chimp
liftOver /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_humanPeakslifted_reverse.bed /project2/gilad/briana/genome_anotation_data/comp_genomes/liftover/hg38ToPanTro5.over.chain.gz /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_humanPeakslifted_reverse.finalCcords.bed /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_humanPeaksunlifted.reverse.final.bed
#chimp to human back to chimp- final lift to human
liftOver /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_chimpPeaks.lifted.reverse.bed /project2/gilad/briana/genome_anotation_data/comp_genomes/liftover/panTro5ToHg38.over.chain.gz /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_chimpPeaks.lifted.reverse.finalHcords.bed /project2/gilad/briana/comparitive_threeprime/data/liftover/filtered_chimpPeaks.unlifted.reverse.final.bed
-change min percentage
The final lifted files have 350111 peaks that lifted from chimp to human and 442100 peaks that lifter human to chimp. I next will find the intersection of these files for the final list. In order to do this I need to create files that have the coordinates in human and in chimp. I can do this using the reverse file and final file.
Process results
human peaks
library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionlibrary(workflowr)This is workflowr version 1.1.1
Run ?workflowr for help getting startedlibrary(tidyverse)── Attaching packages ─────────────────────────────────────────────────── tidyverse 1.2.1 ──✔ ggplot2 3.0.0 ✔ readr 1.1.1
✔ tibble 1.4.2 ✔ purrr 0.2.5
✔ tidyr 0.8.1 ✔ stringr 1.3.1
✔ ggplot2 3.0.0 ✔ forcats 0.3.0── Conflicts ────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()library(reshape2)
Attaching package: 'reshape2'The following object is masked from 'package:tidyr':
smiths#human rev is in human coordinates
human_rev= read.table("../data/liftover/filtered_humanPeakslifted_reverse.bed", col.names = c("human_chr", "human_start", "human_end", "name"), stringsAsFactors = F)
#final coords are in chimp coordinates
human_lifted=read.table("../data/liftover/filtered_humanPeakslifted_reverse.finalCcords.bed", col.names = c("chimp_chr", "chimp_start", "chimp_end", "name"), stringsAsFactors = F)I want to join these files by the name of the peaks keeping only the peaks that are in the final lifted.
human_final=human_lifted %>% left_join(human_rev, by="name")Chimp peaks
#chimp rev in chimp cords
chimp_rev=read.table("../data/liftover/filtered_chimpPeaks.lifted.reverse.bed", col.names = c("chimp_chr", "chimp_start", "chimp_end", "name"), stringsAsFactors = F)
#final chimp lift is in human coords
chimp_lifted=read.table("../data/liftover/filtered_chimpPeaks.lifted.reverse.finalHcords.bed", col.names=c( "human_chr", "human_start", "human_end", "name"),stringsAsFactors = F )Join the files
chimp_final=chimp_lifted %>% left_join(chimp_rev, by="name")Intersection of reciprocal liftover
union_peaks=human_final %>% inner_join(chimp_final, by=c("human_chr", "human_start", "human_end", "chimp_chr", "chimp_start", "chimp_end" ))
peak_names=union_peaks %>% select(name.x, name.y)
colnames(peak_names)= c("human", "chimp")This leaves 76207
I can then seperate these and write out the bedfile. With that I can look at metrics such as how many per gene or distance to last exon.
human_ortho= union_peaks %>% select(human_chr, human_start, human_end, name.x)
chimp_ortho= union_peaks %>% select(chimp_chr, chimp_start, chimp_end, name.y)Write these:
write.table(human_ortho, file="../data/liftover/humanOrthoPeaks.bed", quote = F, row.names = F, col.names = F,sep="\t")
write.table(chimp_ortho, file="../data/liftover/chimpOrthoPeaks.bed", quote= F, row.names = F, col.names = F, sep="\t")
write.table(peak_names, file="../data/liftover/HumanChimpPeaknames.txt", quote=F, row.names = F, col.names = T, sep="\t")Plot results:
I want to plot the number of peaks by proportion that are conserved similar to figure 1a in wang et al. 2018.
hTotal=456566
cTotal= 360860
hConprop= hTotal- 76207
cConprop=cTotal- 76207
hUnConprop=hTotal - hConprop
cUnConprop=cTotal- cConprop
hum=c(hConprop,hUnConprop)
chi=c(cConprop, cUnConprop)
both=as.data.frame(rbind(hum, chi))
colnames(both)=c("Conserved", "Unconserved")
both=both %>% mutate( Species=c("Human", "Chimp"))
both_melt=melt(both, id.vars="Species")
pas_plot=ggplot(both_melt, aes(x=Species, fill=variable, y=value)) + geom_bar(stat="identity", position = "fill") + labs(y="Proportion PAS Peaks", title="Conserved vs Unconserved PAS") + scale_fill_discrete(name="Peak Class") + annotate("text", x=1, y=.85, label=paste("Total PAS:", hTotal, sep=" "))+ annotate("text", x=2, y=.85, label=paste("Total PAS:", cTotal, sep=" ")) + annotate("text", x=1, y=.15, label=paste("Conserved PAS:", "76207", sep=" ")) + annotate("text", x=2, y=.1, label=paste("Conserved PAS:", "76207", sep=" "))
pas_plot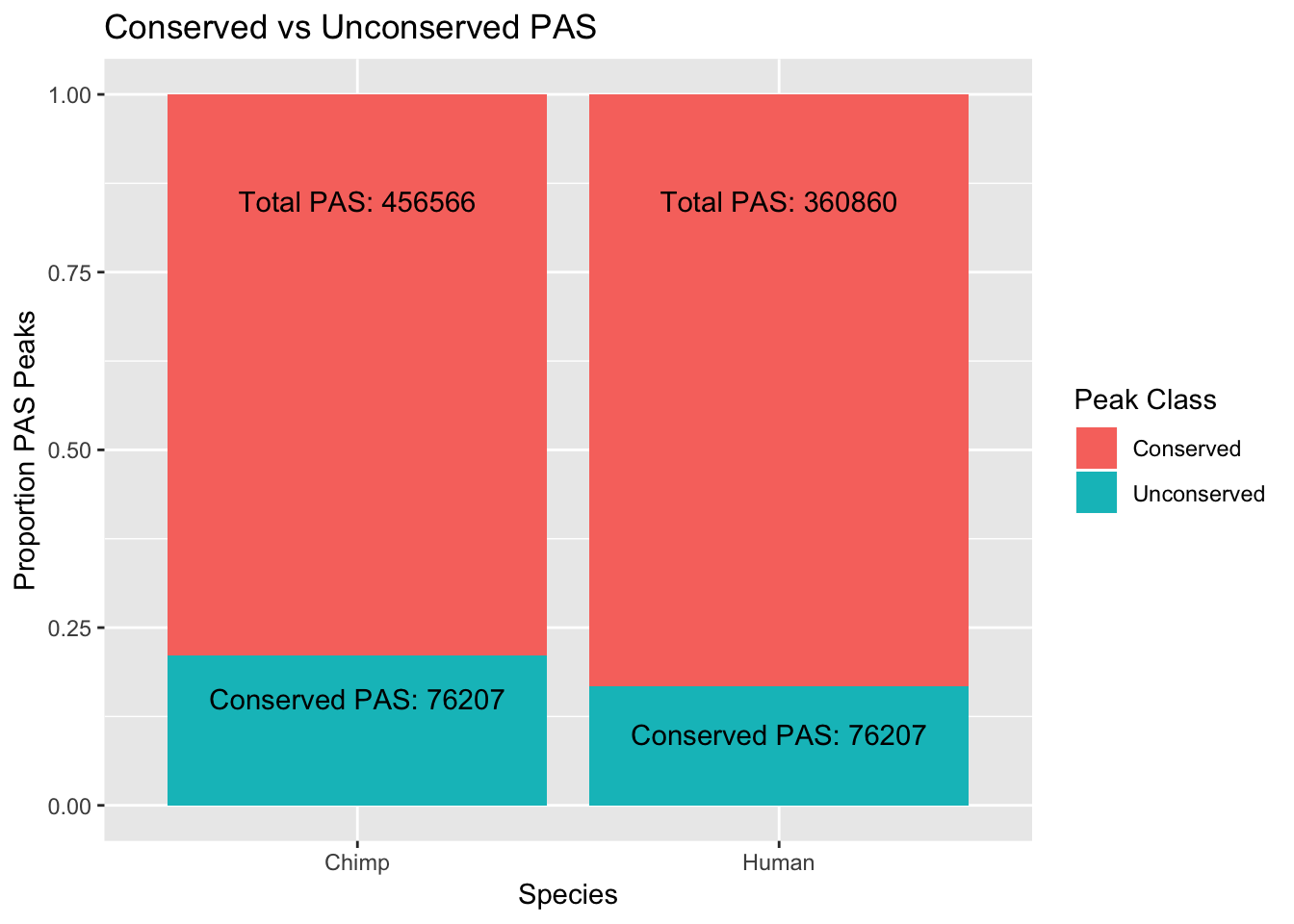
Session information
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Sierra 10.12.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] bindrcpp_0.2.2 reshape2_1.4.3 forcats_0.3.0 stringr_1.3.1
[5] purrr_0.2.5 readr_1.1.1 tidyr_0.8.1 tibble_1.4.2
[9] ggplot2_3.0.0 tidyverse_1.2.1 workflowr_1.1.1 dplyr_0.7.6
loaded via a namespace (and not attached):
[1] tidyselect_0.2.4 haven_1.1.2 lattice_0.20-35
[4] colorspace_1.3-2 htmltools_0.3.6 yaml_2.2.0
[7] rlang_0.2.2 R.oo_1.22.0 pillar_1.3.0
[10] glue_1.3.0 withr_2.1.2 R.utils_2.7.0
[13] modelr_0.1.2 readxl_1.1.0 bindr_0.1.1
[16] plyr_1.8.4 munsell_0.5.0 gtable_0.2.0
[19] cellranger_1.1.0 rvest_0.3.2 R.methodsS3_1.7.1
[22] evaluate_0.11 labeling_0.3 knitr_1.20
[25] broom_0.5.0 Rcpp_0.12.18 scales_1.0.0
[28] backports_1.1.2 jsonlite_1.5 hms_0.4.2
[31] digest_0.6.16 stringi_1.2.4 grid_3.5.1
[34] rprojroot_1.3-2 cli_1.0.0 tools_3.5.1
[37] magrittr_1.5 lazyeval_0.2.1 crayon_1.3.4
[40] whisker_0.3-2 pkgconfig_2.0.2 xml2_1.2.0
[43] lubridate_1.7.4 assertthat_0.2.0 rmarkdown_1.10
[46] httr_1.3.1 rstudioapi_0.7 R6_2.2.2
[49] nlme_3.1-137 git2r_0.23.0 compiler_3.5.1 This reproducible R Markdown analysis was created with workflowr 1.1.1