Overlap with Full GWAS Catelog
Briana Mittleman
3/8/2019
Last updated: 2019-03-11
Checks: 6 0
Knit directory: threeprimeseq/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.2.0). The Report tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(12345) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/.DS_Store
Ignored: data/perm_QTL_trans_noMP_5percov/
Ignored: output/.DS_Store
Untracked files:
Untracked: KalistoAbundance18486.txt
Untracked: analysis/4suDataIGV.Rmd
Untracked: analysis/DirectionapaQTL.Rmd
Untracked: analysis/EmpDistforOverlaps.Rmd
Untracked: analysis/EvaleQTLs.Rmd
Untracked: analysis/NuclearSpecQTL.Rmd
Untracked: analysis/YL_QTL_test.Rmd
Untracked: analysis/groSeqAnalysis.Rmd
Untracked: analysis/ncbiRefSeq_sm.sort.mRNA.bed
Untracked: analysis/snake.config.notes.Rmd
Untracked: analysis/verifyBAM.Rmd
Untracked: analysis/verifybam_dubs.Rmd
Untracked: code/PeaksToCoverPerReads.py
Untracked: code/strober_pc_pve_heatmap_func.R
Untracked: data/18486.genecov.txt
Untracked: data/APApeaksYL.total.inbrain.bed
Untracked: data/AllPeak_counts/
Untracked: data/ApaQTLs/
Untracked: data/ApaQTLs_otherPhen/
Untracked: data/ChromHmmOverlap/
Untracked: data/DistTXN2Peak_genelocAnno/
Untracked: data/FeatureoverlapPeaks/
Untracked: data/GM12878.chromHMM.bed
Untracked: data/GM12878.chromHMM.txt
Untracked: data/GWAS_overlap/
Untracked: data/LianoglouLCL/
Untracked: data/LocusZoom/
Untracked: data/LocusZoom_Unexp/
Untracked: data/LocusZoom_proc/
Untracked: data/MatchedSnps/
Untracked: data/NucSpecQTL/
Untracked: data/NuclearApaQTLs.txt
Untracked: data/PeakCounts/
Untracked: data/PeakCounts_noMP_5perc/
Untracked: data/PeakCounts_noMP_genelocanno/
Untracked: data/PeakUsage/
Untracked: data/PeakUsage_noMP/
Untracked: data/PeakUsage_noMP_GeneLocAnno/
Untracked: data/PeaksUsed/
Untracked: data/PeaksUsed_noMP_5percCov/
Untracked: data/PolyA_DB/
Untracked: data/QTL_overlap/
Untracked: data/RNAkalisto/
Untracked: data/RefSeq_annotations/
Untracked: data/Replicates_usage/
Untracked: data/Signal_Loc/
Untracked: data/TotalApaQTLs.txt
Untracked: data/Totalpeaks_filtered_clean.bed
Untracked: data/UnderstandPeaksQC/
Untracked: data/WASP_STAT/
Untracked: data/YL-SP-18486-T-combined-genecov.txt
Untracked: data/YL-SP-18486-T_S9_R1_001-genecov.txt
Untracked: data/YL_QTL_test/
Untracked: data/apaExamp/
Untracked: data/apaExamp_proc/
Untracked: data/apaQTL_examp_noMP/
Untracked: data/bedgraph_peaks/
Untracked: data/bin200.5.T.nuccov.bed
Untracked: data/bin200.Anuccov.bed
Untracked: data/bin200.nuccov.bed
Untracked: data/clean_peaks/
Untracked: data/comb_map_stats.csv
Untracked: data/comb_map_stats.xlsx
Untracked: data/comb_map_stats_39ind.csv
Untracked: data/combined_reads_mapped_three_prime_seq.csv
Untracked: data/diff_iso_GeneLocAnno/
Untracked: data/diff_iso_proc/
Untracked: data/diff_iso_trans/
Untracked: data/eQTLs_Lietal/
Untracked: data/ensemble_to_genename.txt
Untracked: data/example_gene_peakQuant/
Untracked: data/explainProtVar/
Untracked: data/filtPeakOppstrand_cov_noMP_GeneLocAnno_5perc/
Untracked: data/filtered_APApeaks_merged_allchrom_refseqTrans.closest2End.bed
Untracked: data/filtered_APApeaks_merged_allchrom_refseqTrans.closest2End.noties.bed
Untracked: data/first50lines_closest.txt
Untracked: data/gencov.test.csv
Untracked: data/gencov.test.txt
Untracked: data/gencov_zero.test.csv
Untracked: data/gencov_zero.test.txt
Untracked: data/gene_cov/
Untracked: data/joined
Untracked: data/leafcutter/
Untracked: data/merged_combined_YL-SP-threeprimeseq.bg
Untracked: data/molPheno_noMP/
Untracked: data/mol_overlap/
Untracked: data/mol_pheno/
Untracked: data/nom_QTL/
Untracked: data/nom_QTL_opp/
Untracked: data/nom_QTL_trans/
Untracked: data/nuc6up/
Untracked: data/nuc_10up/
Untracked: data/other_qtls/
Untracked: data/pQTL_otherphen/
Untracked: data/pacbio_cov/
Untracked: data/peakPerRefSeqGene/
Untracked: data/peaks4DT/
Untracked: data/perm_QTL/
Untracked: data/perm_QTL_GeneLocAnno_noMP_5percov/
Untracked: data/perm_QTL_GeneLocAnno_noMP_5percov_3UTR/
Untracked: data/perm_QTL_diffWindow/
Untracked: data/perm_QTL_opp/
Untracked: data/perm_QTL_trans/
Untracked: data/perm_QTL_trans_filt/
Untracked: data/protAndAPAAndExplmRes.Rda
Untracked: data/protAndAPAlmRes.Rda
Untracked: data/protAndExpressionlmRes.Rda
Untracked: data/reads_mapped_three_prime_seq.csv
Untracked: data/smash.cov.results.bed
Untracked: data/smash.cov.results.csv
Untracked: data/smash.cov.results.txt
Untracked: data/smash_testregion/
Untracked: data/ssFC200.cov.bed
Untracked: data/temp.file1
Untracked: data/temp.file2
Untracked: data/temp.gencov.test.txt
Untracked: data/temp.gencov_zero.test.txt
Untracked: data/threePrimeSeqMetaData.csv
Untracked: data/threePrimeSeqMetaData55Ind.txt
Untracked: data/threePrimeSeqMetaData55Ind.xlsx
Untracked: data/threePrimeSeqMetaData55Ind_noDup.txt
Untracked: data/threePrimeSeqMetaData55Ind_noDup.xlsx
Untracked: data/threePrimeSeqMetaData55Ind_noDup_WASPMAP.txt
Untracked: data/threePrimeSeqMetaData55Ind_noDup_WASPMAP.xlsx
Untracked: output/LZ/
Untracked: output/deeptools_plots/
Untracked: output/picard/
Untracked: output/plots/
Untracked: output/qual.fig2.pdf
Unstaged changes:
Modified: analysis/28ind.peak.explore.Rmd
Modified: analysis/CompareLianoglouData.Rmd
Modified: analysis/NewPeakPostMP.Rmd
Modified: analysis/SignalSiteLoc.Rmd
Modified: analysis/apaQTLoverlapGWAS.Rmd
Modified: analysis/characterize_apaQTLs.Rmd
Modified: analysis/cleanupdtseq.internalpriming.Rmd
Modified: analysis/coloc_apaQTLs_protQTLs.Rmd
Modified: analysis/dif.iso.usage.leafcutter.Rmd
Modified: analysis/diff_iso_pipeline.Rmd
Modified: analysis/explainpQTLs.Rmd
Modified: analysis/explore.filters.Rmd
Modified: analysis/fixBWChromNames.Rmd
Modified: analysis/flash2mash.Rmd
Modified: analysis/initialPacBioQuant.Rmd
Modified: analysis/mispriming_approach.Rmd
Modified: analysis/overlapMolQTL.Rmd
Modified: analysis/overlapMolQTL.opposite.Rmd
Modified: analysis/overlap_qtls.Rmd
Modified: analysis/peakOverlap_oppstrand.Rmd
Modified: analysis/peakQCPPlots.Rmd
Modified: analysis/pheno.leaf.comb.Rmd
Modified: analysis/pipeline_55Ind.Rmd
Modified: analysis/swarmPlots_QTLs.Rmd
Modified: analysis/test.max2.Rmd
Modified: analysis/test.smash.Rmd
Modified: analysis/understandPeaks.Rmd
Modified: analysis/unexplainedeQTL_analysis.Rmd
Modified: code/Snakefile
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 1f3e5f6 | Briana Mittleman | 2019-03-11 | add matched snp and result plot |
| html | 9d234b6 | Briana Mittleman | 2019-03-09 | Build site. |
| Rmd | e69f2d3 | Briana Mittleman | 2019-03-09 | add new GWAS overlap |
| html | 55a488c | Briana Mittleman | 2019-03-09 | Build site. |
| Rmd | ea54d47 | Briana Mittleman | 2019-03-09 | add new GWAS overlap |
library(workflowr)This is workflowr version 1.2.0
Run ?workflowr for help getting startedlibrary(tidyverse)── Attaching packages ───────────────────────────────────────────────────────────────────────────────────── tidyverse 1.2.1 ──✔ ggplot2 3.1.0 ✔ purrr 0.3.1
✔ tibble 2.0.1 ✔ dplyr 0.8.0.1
✔ tidyr 0.8.3 ✔ stringr 1.4.0
✔ readr 1.3.1 ✔ forcats 0.4.0 Warning: package 'tibble' was built under R version 3.5.2Warning: package 'tidyr' was built under R version 3.5.2Warning: package 'purrr' was built under R version 3.5.2Warning: package 'dplyr' was built under R version 3.5.2Warning: package 'stringr' was built under R version 3.5.2Warning: package 'forcats' was built under R version 3.5.2── Conflicts ──────────────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()library(data.table)Warning: package 'data.table' was built under R version 3.5.2
Attaching package: 'data.table'The following objects are masked from 'package:dplyr':
between, first, lastThe following object is masked from 'package:purrr':
transposeQTLs
Full GWAS catelog from the table browser. There are 56248699 lines in this file:
/project2/gilad/briana/genome_anotation_data/hg19.GWASCatelog.allsnps
First I want to subset this to a bed file to use. I also want to subset only to SNPs.
Columns: bin chrom chromStart chromEnd name score strand refNCBI refUCSC observed molType class valid avHet avHetSE func locType weight exceptions submitterCount submitters alleleFreqCount alleles alleleNs alleleFreqs bitfields
sed 's/^chr//' /project2/gilad/briana/genome_anotation_data/hg19.GWASCatelog.allsnps > /project2/gilad/briana/genome_anotation_data/hg19.GWASCatelog.allsnps.bedoverlapSNPsGWAS_fixed.py
import pybedtools as pybedtools
def main(infile, outfile):
gwas_file=open("/project2/gilad/briana/genome_anotation_data/hg19.GWASCatelog.allsnps.bed","r")
gwas=pybedtools.BedTool(gwas_file)
snps_file=open(infile, "r")
snps=pybedtools.BedTool(snps_file)
snpOverGWAS=snps.intersect(gwas, wa=True,wb=True)
snpOverGWAS.saveas(outfile)
if __name__ == "__main__":
import sys
import pybedtools
infile=sys.argv[1]
outfile=sys.argv[2]
main(infile, outfile) run_overlapSNPsGWASFixed_proc.sh
#!/bin/bash
#SBATCH --job-name=run_overlapSNPsGWASFixed_proc
#SBATCH --account=pi-yangili1
#SBATCH --time=5:00:00
#SBATCH --output=run_overlapSNPsGWASFixed_proc.out
#SBATCH --error=run_overlapSNPsGWASFixed_proc.err
#SBATCH --partition=broadwl
#SBATCH --mem=10G
#SBATCH --mail-type=END
module load Anaconda3
source activate three-prime-env
python overlapSNPsGWAS_fixed.py "/project2/gilad/briana/threeprimeseq/data/GWAS_overlap_processed/AllOverlapSnps.bed" "/project2/gilad/briana/threeprimeseq/data/GWAS_overlap_processed/GWASoverlapped_AllOverlapSnps.bed"
This analysis gives 9k overlaps with 7k uniq snps. of (53135726 uniq snps)
This makes more sense.
I want to only look at relevent GWAS for LCLs
QTLOverlap=fread("../data/GWAS_overlap/GWASoverlapped_AllOverlapSnps.bed", header=F, col.names = c("chromSnp", "startSnp", "endSnp", "Set", "chromGwas", "startGWAS", "endGWAS", "rsID", "score", "strand"))QTL_overlap_Total=QTLOverlap %>% filter(grepl("Total",Set))
QTL_overlap_Nuclear=QTLOverlap %>% filter(grepl("Nuclear",Set))MAtched snps
I will also need to compare to random snps. (i can use my matched snps/find those in LD)
- /project2/gilad/briana/threeprimeseq/data/MatchedSnp/Nuclear_matched_snps_sort.bed
- /project2/gilad/briana/threeprimeseq/data/MatchedSnp/Total_matched_snps_sort.bed
Switch to format: snp_10_3154947
fixMatchedFormat.py
def fix_format(inbed,outf):
bed=open(inbed, "r")
outF=open(outf, "w")
for ln in bed:
chrom, start, end = ln.split()
outF.write("snp_%s_%s\n"%(chrom, end))
outF.close()
fix_format("/project2/gilad/briana/threeprimeseq/data/MatchedSnp/Nuclear_matched_snps_sort.bed", "/project2/gilad/briana/threeprimeseq/data/MatchedSnp/Nuclear_matched_snps_GEUFormat.txt")
fix_format("/project2/gilad/briana/threeprimeseq/data/MatchedSnp/Total_matched_snps_sort.bed", "/project2/gilad/briana/threeprimeseq/data/MatchedSnp/Total_matched_snps_GEUFormat.txt") subset_plink4Matched_proc.py
def main(genFile, qtlFile, outFile):
#convert snp file to a list:
def file_to_list(file):
snp_list=[]
for ln in file:
snp=ln.strip()
snp_list.append(snp)
return(snp_list)
gen=open(genFile,"r")
fout=open(outFile, "w")
qtls=open(qtlFile, "r")
qtl_list=file_to_list(qtls)
for ln in gen:
snp=ln.split()[2]
if snp in qtl_list:
fout.write(ln)
fout.close()
if __name__ == "__main__":
import sys
chrom=sys.argv[1]
fraction=sys.argv[2]
genFile = "/project2/gilad/briana/threeprimeseq/data/GWAS_overlap/geu_plinkYRI_LDchr%s.ld"%(chrom)
outFile= "/project2/gilad/briana/threeprimeseq/data/GWAS_overlap_matched/%sApaMatch_LD/chr%s.%sMatch.LD.geno.ld"%(fraction,chrom,fraction)
qtlFile= "/project2/gilad/briana/threeprimeseq/data/MatchedSnp/%s_matched_snps_GEUFormat.txt"%(fraction)
main(genFile, qtlFile, outFile) run_subset_plink4Matched_proc.sh
#!/bin/bash
#SBATCH --job-name=run_subset_plink4Matched_proc
#SBATCH --account=pi-yangili1
#SBATCH --time=36:00:00
#SBATCH --output=subset_plink4Matched_proc.out
#SBATCH --error=subset_plink4Matched_proc.err
#SBATCH --partition=broadwl
#SBATCH --mem=30G
#SBATCH --mail-type=END
module load Anaconda3
source activate three-prime-env
for i in {1..22};
do
python subset_plink4Matched_proc.py ${i} "Total"
done
for i in {1..22};
do
python subset_plink4Matched_proc.py ${i} "Nuclear"
doneCat and remove indels:
/project2/gilad/briana/threeprimeseq/data/GWAS_overlap_matched/FractionApaMatch_LD/
cat chr* > allChr.TotalMatch.LD.geno.ld
grep -v indel allChr.TotalMatch.LD.geno.ld > allChr.TotalMatch.LD.geno.ld_noIndel
cat chr* > allChr.NuclearMatch.LD.geno.ld
grep -v indel allChr.NuclearMatch.LD.geno.ld > allChr.NuclearMatch.LD.geno.ld_noIndelmake into bed files:
makeAlloverlapbed_Matched.py
#load files:
QTL_total=open("/project2/gilad/briana/threeprimeseq/data/MatchedSnp/Total_matched_snps_GEUFormat.txt", "r")
QTL_nuclear=open("/project2/gilad/briana/threeprimeseq/data/MatchedSnp/Nuclear_matched_snps_GEUFormat.txt", "r")
LD_total=open("/project2/gilad/briana/threeprimeseq/data/GWAS_overlap_matched/TotalApaMatch_LD/allChr.TotalMatch.LD.geno.ld_noIndel", "r")
LD_nuclear=open("/project2/gilad/briana/threeprimeseq/data/GWAS_overlap_matched/NuclearApaMatch_LD/allChr.NuclearMatch.LD.geno.ld_noIndel", "r")
outFile= open("/project2/gilad/briana/threeprimeseq/data/GWAS_overlap_matched/AllOverlapMatchSnps.bed", "w")
#function for qtl to bed format
def qtl2bed(fqtl, fraction, fout=outFile):
for ln in fqtl:
snp, chrom, pos = ln.split("_")
start=int(pos)-1
end= int(pos)
fout.write("%s\t%d\t%d\tQTL_%s\n"%(chrom, start, end,fraction))
#function for ld to bed format
def ld2bed(fLD, fraction, fout=outFile):
for ln in fLD:
snpID=ln.split()[5]
snp, chrom, pos= snpID.split("_")
start=int(pos)-1
end=int(pos)
fout.write("%s\t%d\t%d\tLD_%s\n"%(chrom, start, end,fraction))
#I will run each of these for both fractions to get all of the snps in the out file.
qtl2bed(QTL_nuclear, "Nuclear")
qtl2bed(QTL_total, "Total")
ld2bed(LD_nuclear, "Nuclear")
ld2bed(LD_total, "Total")
outFile.close()Run the gwas ovelap:
run_overlapSNPsGWASFixed_match.sh
#!/bin/bash
#SBATCH --job-name=run_overlapSNPsGWASFixed_match
#SBATCH --account=pi-yangili1
#SBATCH --time=5:00:00
#SBATCH --output=run_overlapSNPsGWASFixed_match.out
#SBATCH --error=run_overlapSNPsGWASFixed_match.err
#SBATCH --partition=broadwl
#SBATCH --mem=10G
#SBATCH --mail-type=END
module load Anaconda3
source activate three-prime-env
python overlapSNPsGWAS_fixed.py "/project2/gilad/briana/threeprimeseq/data/GWAS_overlap_matched/AllOverlapMatchSnps.bed" "/project2/gilad/briana/threeprimeseq/data/GWAS_overlap_matched/GWASOverlap_AllOverlapMatchSnps.bed"MatchOverlap=fread("../data/GWAS_overlap/GWASOverlap_AllOverlapMatchSnps.bed", header=F, col.names = c("chromSnp", "startSnp", "endSnp", "Set", "chromGwas", "startGWAS", "endGWAS", "rsID", "score", "strand"))MatchOverlap_Total=MatchOverlap %>% filter(grepl("Total",Set))
MatchOverlap_Nuclear=MatchOverlap %>% filter(grepl("Nuclear",Set))Compare:
MatchTot= MatchOverlap_Total %>% select(rsID) %>% unique() %>% nrow()
MatchNuc= MatchOverlap_Nuclear %>% select(rsID) %>% unique() %>% nrow()
QTL_tot=QTL_overlap_Total %>% select(rsID) %>% unique() %>% nrow()
QTL_nuc= QTL_overlap_Nuclear %>% select(rsID) %>% unique() %>% nrow()Number of uniq snps in GWAS catelog
GWAS_snp=53135726
totpval= phyper(QTL_tot, GWAS_snp, GWAS_snp,MatchTot+QTL_tot, lower.tail = F )
nucpval= phyper(QTL_nuc, GWAS_snp, GWAS_snp,MatchNuc+QTL_nuc ,lower.tail = F)GWASdf=as.data.frame(cbind(Fraction=c("Total", "Total", "Nuclear", "Nuclear"),Type=c("QTL", "Match", "QTL", "Match"),Value=c(QTL_tot,MatchTot, QTL_nuc, MatchNuc)))
GWASdf$Value=as.numeric(as.character(GWASdf$Value))
anno_df=data.frame(Type="QTL",Value=6300,
Fraction = factor("Nuclear",levels = c("Nuclear","Total")))
anno_df2=data.frame(Type="QTL",Value=6000,
Fraction = factor("Nuclear",levels = c("Nuclear","Total")))
GwasOverlap=ggplot(GWASdf, aes(x=Type, by=Type, fill=Type, y=Value)) + geom_bar(position="dodge", stat="identity") + facet_grid(~Fraction) + labs(y="N overlap snps",title="apaQTLs overlap with GWAS catelog")+ geom_text(data = anno_df,aes(label="P < .0001")) + scale_fill_manual(values= c("Grey", "Blue")) + geom_text(data = anno_df2,aes(label="***"))
GwasOverlap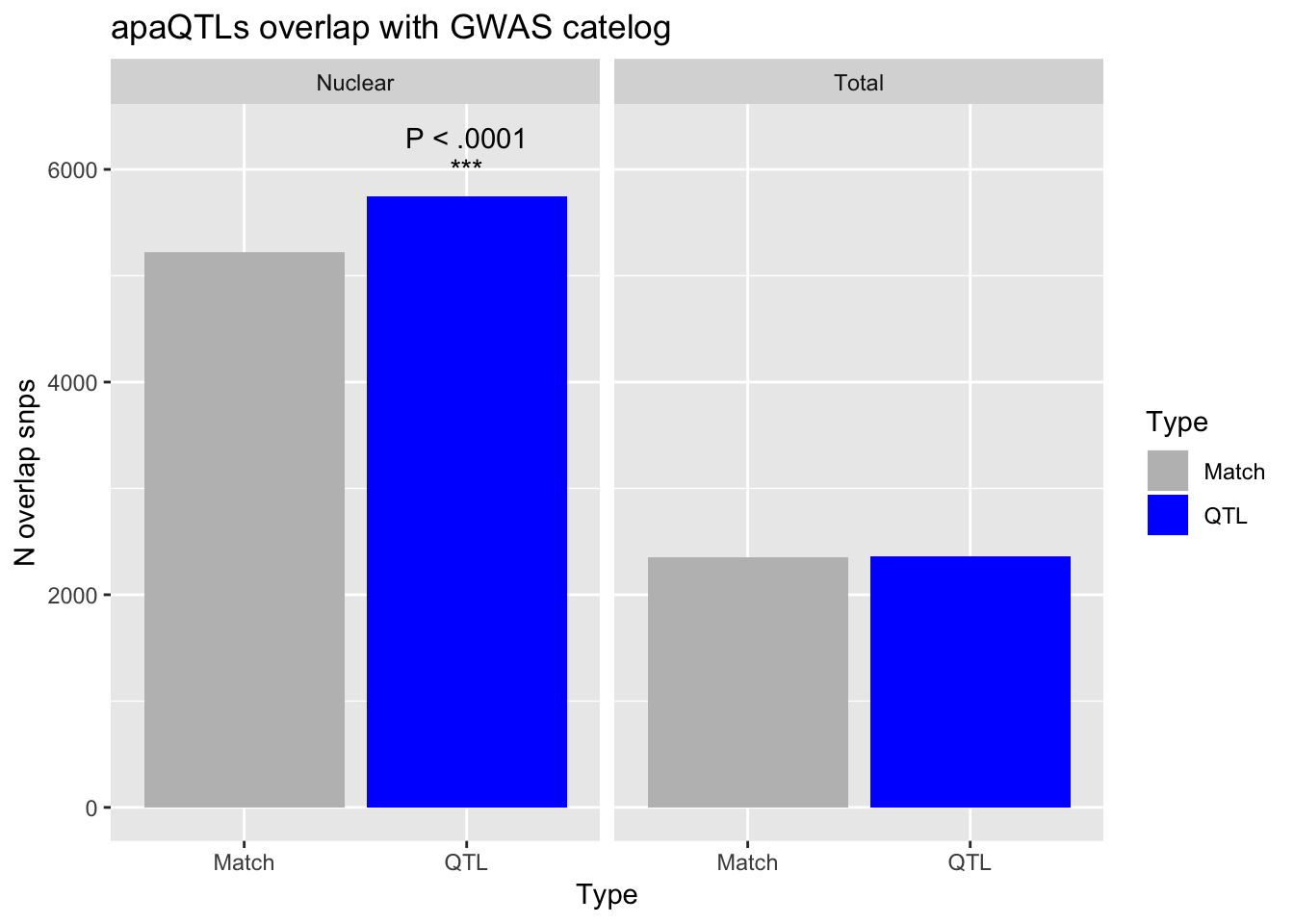
ggsave(GwasOverlap, file="../output/plots/apaQTLsoverlapGWASCatelog.png")Saving 7 x 5 in image
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.14.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] data.table_1.12.0 forcats_0.4.0 stringr_1.4.0
[4] dplyr_0.8.0.1 purrr_0.3.1 readr_1.3.1
[7] tidyr_0.8.3 tibble_2.0.1 ggplot2_3.1.0
[10] tidyverse_1.2.1 workflowr_1.2.0
loaded via a namespace (and not attached):
[1] Rcpp_1.0.0 cellranger_1.1.0 plyr_1.8.4 pillar_1.3.1
[5] compiler_3.5.1 git2r_0.24.0 tools_3.5.1 digest_0.6.18
[9] lubridate_1.7.4 jsonlite_1.6 evaluate_0.13 nlme_3.1-137
[13] gtable_0.2.0 lattice_0.20-38 pkgconfig_2.0.2 rlang_0.3.1
[17] cli_1.0.1 rstudioapi_0.9.0 yaml_2.2.0 haven_2.1.0
[21] xfun_0.5 withr_2.1.2 xml2_1.2.0 httr_1.4.0
[25] knitr_1.21 hms_0.4.2 generics_0.0.2 fs_1.2.6
[29] rprojroot_1.3-2 grid_3.5.1 tidyselect_0.2.5 glue_1.3.0
[33] R6_2.4.0 readxl_1.3.0 rmarkdown_1.11 reshape2_1.4.3
[37] modelr_0.1.4 magrittr_1.5 whisker_0.3-2 backports_1.1.3
[41] scales_1.0.0 htmltools_0.3.6 rvest_0.3.2 assertthat_0.2.0
[45] colorspace_1.4-0 labeling_0.3 stringi_1.3.1 lazyeval_0.2.1
[49] munsell_0.5.0 broom_0.5.1 crayon_1.3.4